bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7578_orf1
Length=73
Score E
Sequences producing significant alignments: (Bits) Value
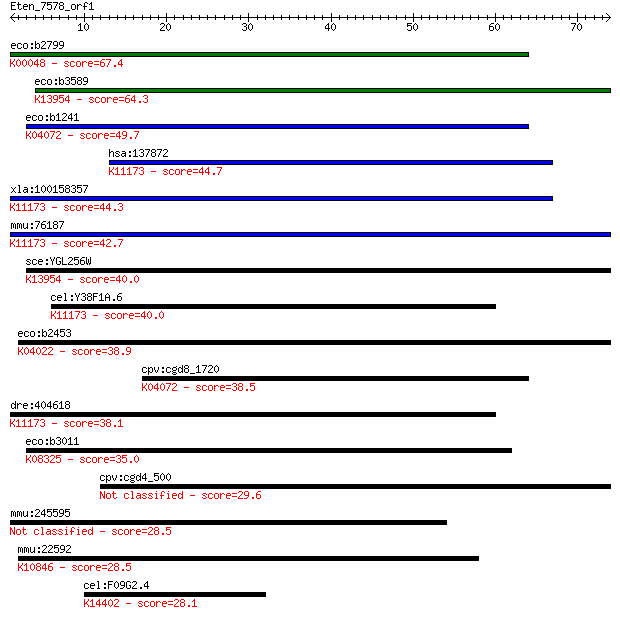
eco:b2799 fucO, ECK2794, JW2770; L-1,2-propanediol oxidoreduct... 67.4 1e-11
eco:b3589 yiaY, ECK3578, JW5648; predicted Fe-containing alcoh... 64.3 1e-10
eco:b1241 adhE, adhC, ana, ECK1235, JW1228; fused acetaldehyde... 49.7 2e-06
hsa:137872 ADHFE1, ADH8, FLJ32430, HOT, MGC48605; alcohol dehy... 44.7 7e-05
xla:100158357 adhfe1; alcohol dehydrogenase, iron containing, ... 44.3 9e-05
mmu:76187 Adhfe1, 6330565B14Rik, AI043035, Adh8; alcohol dehyd... 42.7 3e-04
sce:YGL256W ADH4, NRC465, ZRG5; Adh4p (EC:1.1.1.190 1.1.1.1); ... 40.0 0.002
cel:Y38F1A.6 hypothetical protein; K11173 hydroxyacid-oxoacid ... 40.0 0.002
eco:b2453 eutG, ECK2448, JW2437, yffV; ethanol dehydrogenase i... 38.9 0.004
cpv:cgd8_1720 acetaldehyde reductase plus alcohol dehydrogenas... 38.5 0.005
dre:404618 adhfe1, MGC77479, zgc:77479; alcohol dehydrogenase,... 38.1 0.007
eco:b3011 yqhD, ECK3003, JW2978; aldehyde reductase, NADPH-dep... 35.0 0.065
cpv:cgd4_500 oocyst wall protein 7 29.6
mmu:245595 Zfp711, 2810409C01Rik, A230078I01Rik, Znf711; zinc ... 28.5 4.7
mmu:22592 Ercc5, MGC176031, Xpg; excision repair cross-complem... 28.5 5.3
cel:F09G2.4 cpsf-2; Cleavage and Polyadenylation Specificity F... 28.1 7.8
> eco:b2799 fucO, ECK2794, JW2770; L-1,2-propanediol oxidoreductase
(EC:1.1.1.77); K00048 lactaldehyde reductase [EC:1.1.1.77]
Length=383
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 45/63 (71%), Gaps = 0/63 (0%)
Query 1 VAKVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAI 60
VAKVT+ +DA + + +YDG+ PT +V EG+ ++Q + D+LI +GGGSP D+ KAI
Sbjct 48 VAKVTDKMDAAGLAWAIYDGVVPNPTITVVKEGLGVFQNSGADYLIAIGGGSPQDTCKAI 107
Query 61 GVM 63
G++
Sbjct 108 GII 110
> eco:b3589 yiaY, ECK3578, JW5648; predicted Fe-containing alcohol
dehydrogenase, Pfam00465 family; K13954 alcohol dehydrogenase
[EC:1.1.1.1]
Length=383
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/70 (50%), Positives = 44/70 (62%), Gaps = 1/70 (1%)
Query 4 VTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAIGVM 63
V AL+ NI V+YDG PT + VA G+KL +EN CD +I LGGGSP D AK I +
Sbjct 51 VQKALEERNIFSVIYDGTQPNPTTENVAAGLKLLKENNCDSVISLGGGSPHDCAKGIA-L 109
Query 64 VASDAKKISD 73
VA++ I D
Sbjct 110 VAANGGDIRD 119
> eco:b1241 adhE, adhC, ana, ECK1235, JW1228; fused acetaldehyde-CoA
dehydrogenase/iron-dependent alcohol dehydrogenase/pyruvate-formate
lyase deactivase (EC:1.2.1.10 1.1.1.1); K04072
acetaldehyde dehydrogenase / alcohol dehydrogenase [EC:1.2.1.10
1.1.1.1]
Length=891
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 38/61 (62%), Gaps = 0/61 (0%)
Query 3 KVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAIGV 62
++T+ L A +E V+ + +PT +V +G +L + D +I LGGGSPMD+AK + V
Sbjct 498 QITSVLKAAGVETEVFFEVEADPTLSIVRKGAELANSFKPDVIIALGGGSPMDAAKIMWV 557
Query 63 M 63
M
Sbjct 558 M 558
> hsa:137872 ADHFE1, ADH8, FLJ32430, HOT, MGC48605; alcohol dehydrogenase,
iron containing, 1 (EC:1.1.99.24); K11173 hydroxyacid-oxoacid
transhydrogenase [EC:1.1.99.24]
Length=467
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 13 IEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAIGVMVAS 66
I + VYD + EPTD E ++ Q+ D + +GGGS MD+ KA + +S
Sbjct 102 IPFTVYDNVRVEPTDSSFMEAIEFAQKGAFDAYVAVGGGSTMDTCKAANLYASS 155
> xla:100158357 adhfe1; alcohol dehydrogenase, iron containing,
1 (EC:1.1.99.24); K11173 hydroxyacid-oxoacid transhydrogenase
[EC:1.1.99.24]
Length=463
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 1 VAKVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAI 60
V V N+L I + +YD + EPTDK + ++ ++ + D + +GGGS +D+ KA
Sbjct 86 VKAVLNSLVKNGINFKLYDSVRVEPTDKSFMDAIEFAKKGQFDAFVAVGGGSVIDTCKAA 145
Query 61 GVMVAS 66
+ +S
Sbjct 146 NLYSSS 151
> mmu:76187 Adhfe1, 6330565B14Rik, AI043035, Adh8; alcohol dehydrogenase,
iron containing, 1 (EC:1.1.99.24); K11173 hydroxyacid-oxoacid
transhydrogenase [EC:1.1.99.24]
Length=465
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 37/73 (50%), Gaps = 0/73 (0%)
Query 1 VAKVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAI 60
V V ++L I + VYD + EPTD + ++ ++ D + +GGGS MD+ KA
Sbjct 88 VQIVMDSLSKNGISFQVYDDVRVEPTDGSFMDAIEFAKKGAFDAYVAVGGGSTMDTCKAA 147
Query 61 GVMVASDAKKISD 73
+ +S + D
Sbjct 148 NLYASSPHSEFLD 160
> sce:YGL256W ADH4, NRC465, ZRG5; Adh4p (EC:1.1.1.190 1.1.1.1);
K13954 alcohol dehydrogenase [EC:1.1.1.1]
Length=382
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 39/71 (54%), Gaps = 1/71 (1%)
Query 3 KVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAIGV 62
+V L+ ++ +YD P V G+K+ +E + ++ +GGGS D+AKAI
Sbjct 51 RVQKMLEERDLNVAIYDKTQPNPNIANVTAGLKVLKEQNSEIVVSIGGGSAHDNAKAIA- 109
Query 63 MVASDAKKISD 73
++A++ +I D
Sbjct 110 LLATNGGEIGD 120
> cel:Y38F1A.6 hypothetical protein; K11173 hydroxyacid-oxoacid
transhydrogenase [EC:1.1.99.24]
Length=465
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 6 NALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKA 59
AL NIEY V+D + EPT + + + + + D I +GGGS +D+ KA
Sbjct 92 QALKMVNIEYEVFDDVLIEPTVNSMQKAIAFAKSKQFDSFIAVGGGSVIDTTKA 145
> eco:b2453 eutG, ECK2448, JW2437, yffV; ethanol dehydrogenase
involved in ethanolamine utilization; aldehyde reductase, converts
acetaldehyde to ethanol; K04022 alcohol dehydrogenase
Length=395
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 38/72 (52%), Gaps = 0/72 (0%)
Query 2 AKVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAIG 61
A +T +L I ++ GEP V V +E+ CD +I GGGS +D+AKA+
Sbjct 67 AGLTRSLTVKGIAMTLWPCPVGEPCITDVCAAVAQLRESGCDGVIAFGGGSVLDAAKAVT 126
Query 62 VMVASDAKKISD 73
++V + +++
Sbjct 127 LLVTNPDSTLAE 138
> cpv:cgd8_1720 acetaldehyde reductase plus alcohol dehydrogenase
(AdhE) of possible bacterial origin ; K04072 acetaldehyde
dehydrogenase / alcohol dehydrogenase [EC:1.2.1.10 1.1.1.1]
Length=867
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 17 VYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAIGVM 63
++ + EP + V E VK ++ D LIG GGGSPMD++K + +M
Sbjct 503 MFAEVPPEPDVETVKEIVKRLNVSKPDCLIGFGGGSPMDASKLVRLM 549
> dre:404618 adhfe1, MGC77479, zgc:77479; alcohol dehydrogenase,
iron containing, 1 (EC:1.1.1.1); K11173 hydroxyacid-oxoacid
transhydrogenase [EC:1.1.99.24]
Length=471
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 1 VAKVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKA 59
VA V ++L +++ +Y+ + EPTDK + ++ D + +GGGS +D+ KA
Sbjct 94 VAAVLDSLIKHGVKHKLYEDVRVEPTDKSFKAAIDFAKKGHFDVYVAVGGGSVIDTCKA 152
> eco:b3011 yqhD, ECK3003, JW2978; aldehyde reductase, NADPH-dependent;
K08325 NADP-dependent alcohol dehydrogenase [EC:1.1.-.-]
Length=387
Score = 35.0 bits (79), Expect = 0.065, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 2/59 (3%)
Query 3 KVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAIG 61
+V +AL ++ + + GI P + + VKL +E + FL+ +GGGS +D K I
Sbjct 49 QVLDALKGMDV--LEFGGIEPNPAYETLMNAVKLVREQKVTFLLAVGGGSVLDGTKFIA 105
> cpv:cgd4_500 oocyst wall protein 7
Length=840
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 27/62 (43%), Gaps = 8/62 (12%)
Query 12 NIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSAKAIGVMVASDAKKI 71
N E DGIS P K ++ DF++G G + K GV+ A DA+K
Sbjct 132 NFEVPTIDGISSSPQQK-------CFKMEVADFVLGCPNGWAFNGIKCTGVLTA-DAQKA 183
Query 72 SD 73
D
Sbjct 184 CD 185
> mmu:245595 Zfp711, 2810409C01Rik, A230078I01Rik, Znf711; zinc
finger protein 711
Length=805
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 2/53 (3%)
Query 1 VAKVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSP 53
VA + + D ++E+VV D +SG + MV+E V L ++ + +I GGG P
Sbjct 130 VADLVSGPDG-HLEHVVQDCVSGVDSPTMVSEEV-LVTNSDTETVIQAGGGVP 180
> mmu:22592 Ercc5, MGC176031, Xpg; excision repair cross-complementing
rodent repair deficiency, complementation group 5;
K10846 DNA excision repair protein ERCC-5
Length=1170
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 22/56 (39%), Positives = 27/56 (48%), Gaps = 3/56 (5%)
Query 2 AKVTNALDACNIEYVVYDGISGEPTDKMVAEGVKLYQENECDFLIGLGGGSPMDSA 57
A + ALD N E V G S + +KM+ G L QE D GGG P D+A
Sbjct 389 AAIQKALDDDNDEKV--SGSSDDLAEKMLL-GSGLEQEEHADETAERGGGVPFDTA 441
> cel:F09G2.4 cpsf-2; Cleavage and Polyadenylation Specificity
Factor family member (cpsf-2); K14402 cleavage and polyadenylation
specificity factor subunit 2
Length=843
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 12/23 (52%), Positives = 18/23 (78%), Gaps = 1/23 (4%)
Query 10 ACNIEYVVYDGIS-GEPTDKMVA 31
+C IE++ Y+GIS GE T K++A
Sbjct 566 SCRIEFIEYEGISDGESTKKLLA 588
Lambda K H
0.313 0.133 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2011623444
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40