bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7576_orf1
Length=192
Score E
Sequences producing significant alignments: (Bits) Value
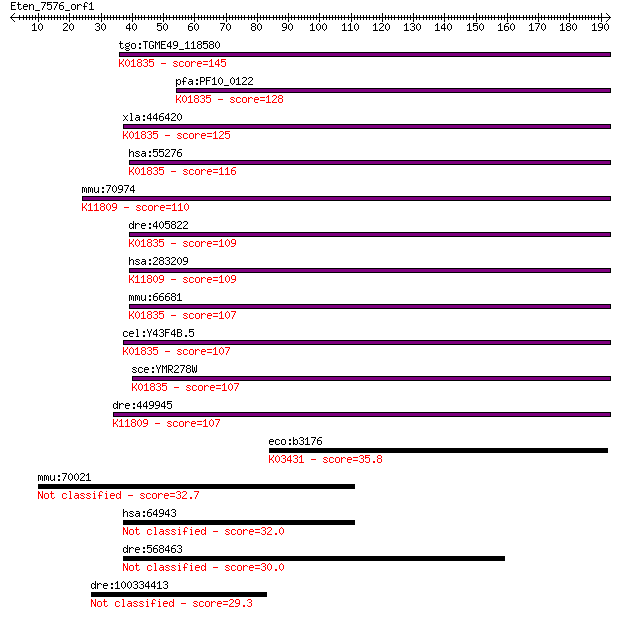
tgo:TGME49_118580 phosphoglucomutase, putative (EC:5.4.2.8); K... 145 7e-35
pfa:PF10_0122 phosphoglucomutase, putative; K01835 phosphogluc... 128 1e-29
xla:446420 pgm2l1, MGC83889; phosphoglucomutase 2-like 1 (EC:2... 125 1e-28
hsa:55276 PGM2, FLJ10983; phosphoglucomutase 2 (EC:5.4.2.2 5.4... 116 5e-26
mmu:70974 Pgm2l1, 4931406N15Rik, AI553438, BM32A; phosphogluco... 110 3e-24
dre:405822 pgm2, MGC77113, zgc:77113; phosphoglucomutase 2 (EC... 109 6e-24
hsa:283209 PGM2L1, BM32A, FLJ32029; phosphoglucomutase 2-like ... 109 6e-24
mmu:66681 Pgm1, 3230402E02Rik, Pgm-1, Pgm2; phosphoglucomutase... 107 2e-23
cel:Y43F4B.5 hypothetical protein; K01835 phosphoglucomutase [... 107 2e-23
sce:YMR278W PGM3; Pgm3p; K01835 phosphoglucomutase [EC:5.4.2.2] 107 4e-23
dre:449945 pgm2l1, im:7140576, zgc:198285; phosphoglucomutase ... 107 4e-23
eco:b3176 glmM, ECK3165, JW3143, mrsA, yhbF; phosphoglucosamin... 35.8 0.082
mmu:70021 Nt5dc2, 2510015F01Rik; 5'-nucleotidase domain contai... 32.7 0.86
hsa:64943 NT5DC2, FLJ12442; 5'-nucleotidase domain containing ... 32.0 1.3
dre:568463 ryr2b; ryanodine receptor 2b (cardiac) 30.0 5.0
dre:100334413 enzymatic polyprotein; Endonuclease; Reverse tra... 29.3 7.8
> tgo:TGME49_118580 phosphoglucomutase, putative (EC:5.4.2.8);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=597
Score = 145 bits (367), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 73/157 (46%), Positives = 98/157 (62%), Gaps = 1/157 (0%)
Query 36 RLSDSHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLRGLMGP 95
+ + L+A W + DR +T + +LL+ E+EL ++F L FGTAGLRG MG
Sbjct 7 KFVNGALEAAVNFWRSVDRREETQKETLELLKNLTEDELAKLFLARLEFGTAGLRGRMGA 66
Query 96 GYSRLNDVTIQQASQGLAMYLLDRHGKEACAARGIVIGRDCRHNSERFANLAAATFLSVG 155
G+SR+NDVTIQQ +QG +L+D G++ RG+VIG D RHNS RFA L AA FLS G
Sbjct 67 GFSRMNDVTIQQTTQGYCAFLVDVFGEDG-KDRGVVIGFDARHNSRRFAQLTAAVFLSKG 125
Query 156 FKVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNPK 192
F+V L V TP+V + + +A +TASHNPK
Sbjct 126 FRVQLFSDIVHTPMVPYTVVAANCIAGIMITASHNPK 162
> pfa:PF10_0122 phosphoglucomutase, putative; K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=593
Score = 128 bits (321), Expect = 1e-29, Method: Composition-based stats.
Identities = 65/140 (46%), Positives = 93/140 (66%), Gaps = 1/140 (0%)
Query 54 RNPQTLA-QAQQLLREAPEEELKEIFCKHLVFGTAGLRGLMGPGYSRLNDVTIQQASQGL 112
+ PQ L + ++L++ EEELK +F K L FGTAGLRG M G++ +N VTI Q +QGL
Sbjct 20 KKPQYLIDETIEILKKNDEEELKCLFLKRLNFGTAGLRGKMCVGFNAMNVVTIMQTTQGL 79
Query 113 AMYLLDRHGKEACAARGIVIGRDCRHNSERFANLAAATFLSVGFKVLLCGAPVATPLVAF 172
YL++ +G C RGI+ G D R++SE FA++AA+ LS GF+V L VATP++ +
Sbjct 80 CSYLINTYGLNLCKNRGIIFGFDGRYHSESFAHVAASVCLSKGFRVYLFAQTVATPILCY 139
Query 173 AALQQQTVAAAAVTASHNPK 192
+ L++ + VTASHNPK
Sbjct 140 SNLKKNCLCGVMVTASHNPK 159
> xla:446420 pgm2l1, MGC83889; phosphoglucomutase 2-like 1 (EC:2.7.1.106);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=610
Score = 125 bits (313), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 68/162 (41%), Positives = 90/162 (55%), Gaps = 8/162 (4%)
Query 37 LSDSHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLRGLMGPG 96
+ D+ L W+ D+NP+TLA +Q++ + +EEL+ F + FGTAGLR MGPG
Sbjct 8 VGDARLDKAVTDWIKWDKNPKTLALVKQMVADGKKEELQACFGSRMEFGTAGLRAAMGPG 67
Query 97 YSRLNDVTIQQASQGLAMYLLDRHGKEACAARGIVIGRDCR------HNSERFANLAAAT 150
S++ND+TI Q +QG YL RG+VIG D R +S+RFA LAA T
Sbjct 68 ISQMNDLTIIQTTQGFCRYL--EKNISDLKERGVVIGYDARAHPASGGSSKRFARLAATT 125
Query 151 FLSVGFKVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNPK 192
F+S G KV + TP V +A A VTASHNPK
Sbjct 126 FVSQGIKVYMFSDITPTPFVPYAVTHLNLCAGIMVTASHNPK 167
> hsa:55276 PGM2, FLJ10983; phosphoglucomutase 2 (EC:5.4.2.2 5.4.2.7);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=612
Score = 116 bits (290), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 67/160 (41%), Positives = 86/160 (53%), Gaps = 8/160 (5%)
Query 39 DSHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLRGLMGPGYS 98
D+ L + WL D+N TL ++L+ E +EEL++ F + FGTAGLR MGPG S
Sbjct 12 DARLDQETAQWLRWDKNSLTLEAVKRLIAEGNKEELRKCFGARMEFGTAGLRAAMGPGIS 71
Query 99 RLNDVTIQQASQGLAMYLLDRHGKEACAARGIVIGRDCR------HNSERFANLAAATFL 152
R+ND+TI Q +QG YL + +GIVI D R +S RFA LAA TF+
Sbjct 72 RMNDLTIIQTTQGFCRYLEKQFSD--LKQKGIVISFDARAHPSSGGSSRRFARLAATTFI 129
Query 153 SVGFKVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNPK 192
S G V L TP V F + A +TASHNPK
Sbjct 130 SQGIPVYLFSDITPTPFVPFTVSHLKLCAGIMITASHNPK 169
> mmu:70974 Pgm2l1, 4931406N15Rik, AI553438, BM32A; phosphoglucomutase
2-like 1 (EC:2.7.1.106); K11809 glucose-1,6-bisphosphate
synthase [EC:2.7.1.106]
Length=621
Score = 110 bits (275), Expect = 3e-24, Method: Composition-based stats.
Identities = 67/179 (37%), Positives = 92/179 (51%), Gaps = 18/179 (10%)
Query 24 LHSKMDAESYLNRLSDSHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLV 83
L+S + YL D L WL D+NP+T Q + LLR +EL++ C +
Sbjct 9 LNSNLLHAPYLT--GDPQLDTAIGQWLRWDKNPKTKEQIENLLRNGMNKELRDRLCCRMT 66
Query 84 FGTAGLRGLMGPGYSRLNDVTIQQASQGLAMYLLDRHGKEACAA----RGIVIGRDCRH- 138
FGTAGLR MG G+ +ND+T+ Q++QG+ YL E C + RG V+G D R
Sbjct 67 FGTAGLRSAMGAGFCYINDLTVIQSTQGMYKYL------ERCFSDFKQRGFVVGYDTRGQ 120
Query 139 -----NSERFANLAAATFLSVGFKVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNPK 192
+S+R A L AA L+ V L V TP V +A + + VA +TASHN K
Sbjct 121 VTSSCSSQRLAKLTAAVLLAKDIPVYLFSRYVPTPFVPYAVQELKAVAGVMITASHNRK 179
> dre:405822 pgm2, MGC77113, zgc:77113; phosphoglucomutase 2 (EC:5.4.2.2);
K01835 phosphoglucomutase [EC:5.4.2.2]
Length=611
Score = 109 bits (272), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 68/164 (41%), Positives = 85/164 (51%), Gaps = 16/164 (9%)
Query 39 DSHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLRGLMGPGYS 98
DS L WL D+NP+T A + ++++ EL++ F + FGTAGLR MGPG S
Sbjct 10 DSRLDQAVTQWLQYDKNPKTAAAVRSMVKDGALSELQKCFGARMEFGTAGLRAAMGPGVS 69
Query 99 RLNDVTIQQASQGLAMYLLDRHGKEAC----AARGIVIGRDCR------HNSERFANLAA 148
+ND+TI Q +QG YL E C RG+VIG D R +S+RFA LAA
Sbjct 70 CMNDLTIIQTTQGFCAYL------EECFVDLKQRGVVIGFDARAHPPSGGSSKRFACLAA 123
Query 149 ATFLSVGFKVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNPK 192
+S G V L TP V FA A VTASHNPK
Sbjct 124 CVLMSRGVPVHLFSDITPTPYVPFAVSHLGLCAGIMVTASHNPK 167
> hsa:283209 PGM2L1, BM32A, FLJ32029; phosphoglucomutase 2-like
1 (EC:2.7.1.106); K11809 glucose-1,6-bisphosphate synthase
[EC:2.7.1.106]
Length=622
Score = 109 bits (272), Expect = 6e-24, Method: Composition-based stats.
Identities = 63/164 (38%), Positives = 86/164 (52%), Gaps = 16/164 (9%)
Query 39 DSHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLRGLMGPGYS 98
D L WL D+NP+T Q + LLR +EL++ C + FGTAGLR MG G+
Sbjct 22 DPQLDTAIGQWLRWDKNPKTKEQIENLLRNGMNKELRDRLCCRMTFGTAGLRSAMGAGFC 81
Query 99 RLNDVTIQQASQGLAMYLLDRHGKEACAA----RGIVIGRDCRH------NSERFANLAA 148
+ND+T+ Q++QG+ YL E C + RG V+G D R +S+R A L A
Sbjct 82 YINDLTVIQSTQGMYKYL------ERCFSDFKQRGFVVGYDTRGQVTSSCSSQRLAKLTA 135
Query 149 ATFLSVGFKVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNPK 192
A L+ V L V TP V +A + + VA +TASHN K
Sbjct 136 AVLLAKDVPVYLFSRYVPTPFVPYAVQKLKAVAGVMITASHNRK 179
> mmu:66681 Pgm1, 3230402E02Rik, Pgm-1, Pgm2; phosphoglucomutase
1 (EC:5.4.2.2 5.4.2.7); K01835 phosphoglucomutase [EC:5.4.2.2]
Length=620
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 63/160 (39%), Positives = 83/160 (51%), Gaps = 8/160 (5%)
Query 39 DSHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLRGLMGPGYS 98
D+ L + WL D+NP T +QL+ +EEL++ F + FGTAGLR MG G S
Sbjct 20 DARLDQETAQWLRWDQNPLTSESVKQLIAGGNKEELRKCFGARMEFGTAGLRAPMGAGIS 79
Query 99 RLNDVTIQQASQGLAMYLLDRHGKEACAARGIVIGRDCR------HNSERFANLAAATFL 152
R+ND+TI Q +QG YL + RG+VI D R +S RFA LAA F+
Sbjct 80 RMNDLTIIQTTQGFCRYLEKQFSD--LKQRGVVISFDARAHPASGGSSRRFARLAATAFI 137
Query 153 SVGFKVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNPK 192
+ G V L TP V + + A +TASHNPK
Sbjct 138 TQGVPVYLFSDITPTPFVPYTVSHLKLCAGIMITASHNPK 177
> cel:Y43F4B.5 hypothetical protein; K01835 phosphoglucomutase
[EC:5.4.2.2]
Length=595
Score = 107 bits (268), Expect = 2e-23, Method: Composition-based stats.
Identities = 61/156 (39%), Positives = 87/156 (55%), Gaps = 3/156 (1%)
Query 37 LSDSHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLRGLMGPG 96
L + L Q WLA D+N + + Q+L+ E + LK LVFGTAG+R M G
Sbjct 3 LGCAKLDKQVADWLAWDKNDKNRNEIQKLVDEKNVDALKARMDTRLVFGTAGVRSPMQAG 62
Query 97 YSRLNDVTIQQASQGLAMYLLDRHGKEACAARGIVIGRDCRHNSERFANLAAATFLSVGF 156
+ RLND+TI Q + G A ++L+ +G+ G+ IG D R+NS RFA L+A F+
Sbjct 63 FGRLNDLTIIQITHGFARHMLNVYGQ---PKNGVAIGFDGRYNSRRFAELSANVFVRNNI 119
Query 157 KVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNPK 192
V L TP+V++A ++ A +TASHNPK
Sbjct 120 PVYLFSEVSPTPVVSWATIKLGCDAGLIITASHNPK 155
> sce:YMR278W PGM3; Pgm3p; K01835 phosphoglucomutase [EC:5.4.2.2]
Length=622
Score = 107 bits (266), Expect = 4e-23, Method: Composition-based stats.
Identities = 63/156 (40%), Positives = 83/156 (53%), Gaps = 7/156 (4%)
Query 40 SHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLRGLMGPGYSR 99
S LK W DRNP+T+ + L +++ EL + F + FGTAGLR M G+SR
Sbjct 11 SDLKDPISLWFKQDRNPKTIEEVTALCKKSDWNELHKRFDSRIQFGTAGLRSQMQAGFSR 70
Query 100 LNDVTIQQASQGLAMYLLDRHGKEACAARGIVIGRDCRHNSERFANLAAATFLSVGFKVL 159
+N + + QASQGLA Y+ + A V+G D R +S+ FA AA FL GFKV
Sbjct 71 MNTLVVIQASQGLATYVRQQFPDNLVA----VVGHDHRFHSKEFARATAAAFLLKGFKVH 126
Query 160 LCGAP---VATPLVAFAALQQQTVAAAAVTASHNPK 192
V TPLV FA + + +TASHNPK
Sbjct 127 YLNPDHEFVHTPLVPFAVDKLKASVGVMITASHNPK 162
> dre:449945 pgm2l1, im:7140576, zgc:198285; phosphoglucomutase
2-like 1 (EC:2.7.1.106); K11809 glucose-1,6-bisphosphate synthase
[EC:2.7.1.106]
Length=619
Score = 107 bits (266), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 63/165 (38%), Positives = 83/165 (50%), Gaps = 8/165 (4%)
Query 34 LNRLSDSHLKAQARAWLATDRNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLRGLM 93
L+ D L W+ D+NP T Q + L++E EL+ C + FGTAGLR M
Sbjct 14 LSATGDPVLDKAVSQWMTWDKNPLTREQIESLVQEGRVVELRRRLCSRMTFGTAGLRAAM 73
Query 94 GPGYSRLNDVTIQQASQGLAMYLLDRHGKEACAARGIVIGRDCRH------NSERFANLA 147
G G++R+ND+TI Q++QGL YL RG+V+G D R SER A L
Sbjct 74 GAGFARINDLTIIQSTQGLYKYLAKCFPD--LKTRGLVVGYDTRAQASSGCTSERLAKLT 131
Query 148 AATFLSVGFKVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNPK 192
AA L V L V TP V +A ++ A +TASHN K
Sbjct 132 AAVMLCKDVPVYLFSTYVPTPFVPYAVMKYGAAAGVMITASHNRK 176
> eco:b3176 glmM, ECK3165, JW3143, mrsA, yhbF; phosphoglucosamine
mutase (EC:5.4.2.10); K03431 phosphoglucosamine mutase [EC:5.4.2.10]
Length=445
Score = 35.8 bits (81), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 34/108 (31%), Positives = 50/108 (46%), Gaps = 9/108 (8%)
Query 84 FGTAGLRGLMGPGYSRLNDVTIQQASQGLAMYLLDRHGKEACAARGIVIGRDCRHNSERF 143
FGT G+RG +G + V + G +L RHG +R I+IG+D R +
Sbjct 7 FGTDGIRGRVGDAPITPDFVLKLGWAAG---KVLARHG-----SRKIIIGKDTRISGYML 58
Query 144 ANLAAATFLSVGFKVLLCGAPVATPLVAFAALQQQTVAAAAVTASHNP 191
+ A + G L G P+ TP VA+ + A ++ASHNP
Sbjct 59 ESALEAGLAAAGLSALFTG-PMPTPAVAYLTRTFRAEAGIVISASHNP 105
> mmu:70021 Nt5dc2, 2510015F01Rik; 5'-nucleotidase domain containing
2 (EC:3.1.3.5)
Length=390
Score = 32.7 bits (73), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 47/107 (43%), Gaps = 11/107 (10%)
Query 10 ISDYFAFILHLFCKLHSKMDAESYLNRLSDSHLKAQARAWLATD------RNPQTLAQAQ 63
+ DYF F ++H D + + D H+K W+ D R +T A
Sbjct 69 VVDYFLGHGLEFDQVHLYKDVT---DAIRDVHVKGLMYQWIEQDMEKYILRGDETFAVLS 125
Query 64 QLLREAPEEELKEIFCKHLVFGTAGLRGLMGPGYSRLNDVTIQQASQ 110
+L+ A ++L I F G+R ++GP + +L DV I QA +
Sbjct 126 RLV--AHGKQLFLITNSPFSFVDKGMRHMVGPDWRQLFDVVIVQADK 170
> hsa:64943 NT5DC2, FLJ12442; 5'-nucleotidase domain containing
2 (EC:3.1.3.5)
Length=557
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 37/80 (46%), Gaps = 8/80 (10%)
Query 37 LSDSHLKAQARAWLATD------RNPQTLAQAQQLLREAPEEELKEIFCKHLVFGTAGLR 90
+ D H+K W+ D R +T A +L+ A ++L I F G+R
Sbjct 260 IRDVHVKGLMYQWIEQDMEKYILRGDETFAVLSRLV--AHGKQLFLITNSPFSFVDKGMR 317
Query 91 GLMGPGYSRLNDVTIQQASQ 110
++GP + +L DV I QA +
Sbjct 318 HMVGPDWRQLFDVVIVQADK 337
> dre:568463 ryr2b; ryanodine receptor 2b (cardiac)
Length=4882
Score = 30.0 bits (66), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 33/131 (25%), Positives = 56/131 (42%), Gaps = 27/131 (20%)
Query 37 LSDSHLKAQARAWLATDR-------NPQTLAQAQQLLREAPEEELKEIFCK--HLVFGTA 87
LSD H + + +++ P + Q++ +L +A +EEL E + H ++
Sbjct 947 LSDEHAEENVKYVRLSNKYELWNGYKPAPVDQSRVVLTDA-QEELVEYLAENEHNIWARE 1005
Query 88 GLRGLMGPGYSRLNDVTIQQASQGLAMYLLDRHGKEACAARGIVIGRDCRHNSERFANLA 147
+R G Y DV ++++Q + YLLD K+A GRD + A
Sbjct 1006 RIR--QGWSYGPQQDVKFKRSTQLVPFYLLDERYKQA--------GRDAMRD-------A 1048
Query 148 AATFLSVGFKV 158
T L G+ V
Sbjct 1049 LGTLLGFGYTV 1059
> dre:100334413 enzymatic polyprotein; Endonuclease; Reverse transcriptase,
putative-like
Length=1766
Score = 29.3 bits (64), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 34/67 (50%), Gaps = 11/67 (16%)
Query 27 KMDAESYLNR-----LSDSHLK----AQARAWLATDRNPQTLAQAQQLLREAPEEELKE- 76
KMD + R + DSHL+ A+ARA + D PQ ++ Q L E+EL+E
Sbjct 484 KMDELMMIPRDEVSAIIDSHLEWNIGAKARAMTSIDHLPQVMSVGQDTLPAYTEQELREK 543
Query 77 -IFCKHL 82
+ KHL
Sbjct 544 QLSDKHL 550
Lambda K H
0.325 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5558593832
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40