bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7520_orf2
Length=172
Score E
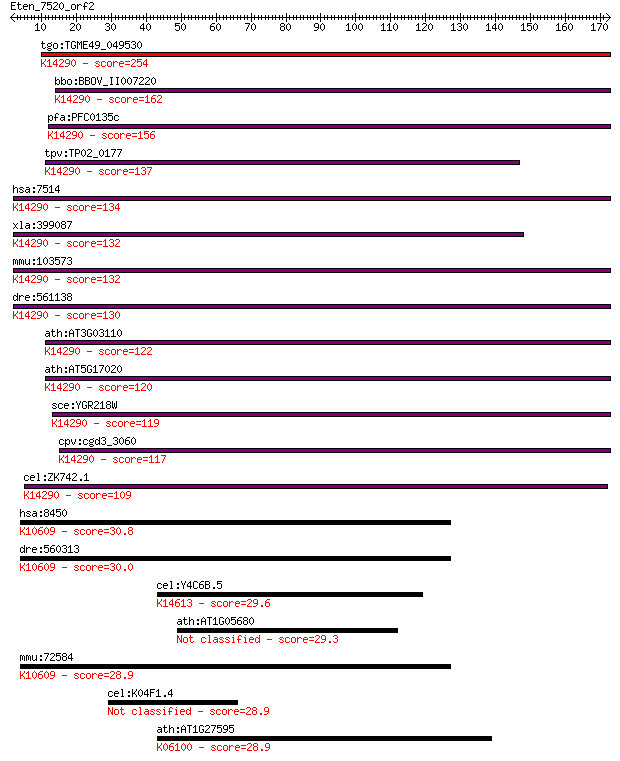
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_049530 exportin, putative ; K14290 exportin-1 254 1e-67
bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1 162 6e-40
pfa:PFC0135c exportin 1, putative; K14290 exportin-1 156 3e-38
tpv:TP02_0177 importin beta-related nuclear transport factor; ... 137 1e-32
hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homo... 134 2e-31
xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog); ... 132 7e-31
mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolo... 132 7e-31
dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b; K1... 130 2e-30
ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14... 122 4e-28
ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transpor... 120 3e-27
sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1 119 7e-27
cpv:cgd3_3060 exportin 1 ; K14290 exportin-1 117 2e-26
cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family m... 109 6e-24
hsa:8450 CUL4B, DKFZp686F1470, KIAA0695, MRXHF2, MRXS15, MRXSC... 30.8 2.1
dre:560313 cul4b, im:7140950, im:7150765; cullin 4B; K10609 cu... 30.0 3.5
cel:Y4C6B.5 hypothetical protein; K14613 MFS transporter, PCFT... 29.6 4.8
ath:AT1G05680 UDP-glucoronosyl/UDP-glucosyl transferase family... 29.3 6.7
mmu:72584 Cul4b, 2700050M05Rik, AA409770, KIAA0695, mKIAA0695;... 28.9 8.0
cel:K04F1.4 srw-91; Serpentine Receptor, class W family member... 28.9 8.4
ath:AT1G27595 hypothetical protein; K06100 symplekin 28.9 9.5
> tgo:TGME49_049530 exportin, putative ; K14290 exportin-1
Length=1125
Score = 254 bits (648), Expect = 1e-67, Method: Compositional matrix adjust.
Identities = 116/163 (71%), Positives = 142/163 (87%), Gaps = 0/163 (0%)
Query 10 RKQLVTDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVIFDYIFD 69
RK++ +LG +LQP+L DY+NN PDTRD EVLTL+SVL +RLD++I+ VLPVIF+++F+
Sbjct 821 RKEIAEKLLGDLLQPVLSDYKNNIPDTRDCEVLTLLSVLTTRLDSNISPVLPVIFEFVFE 880
Query 70 CTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEHPS 129
TL MIK+DFQSYPDHRE+FY LLKA NQHCF GLF+LP+ QLKA+VESLVWAFKHEHPS
Sbjct 881 STLNMIKTDFQSYPDHREKFYELLKACNQHCFDGLFALPAHQLKAYVESLVWAFKHEHPS 940
Query 130 LAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYFSLMKEILLVL 172
+AE+GLQVT+EFL +LI KREVL+DFC +YFSLMKE LLVL
Sbjct 941 VAEQGLQVTYEFLLKLINDKREVLSDFCNLFYFSLMKETLLVL 983
> bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1
Length=1186
Score = 162 bits (410), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 84/172 (48%), Positives = 113/172 (65%), Gaps = 14/172 (8%)
Query 14 VTDMLGQ-VLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVIFDYIFDCTL 72
V +ML Q + +L YRN +TRD+EV+ +++ L+ +L + VLP IF+ IFDCTL
Sbjct 859 VLNMLIQSITSTVLVVYRNCLAETRDHEVICVVTTLIEKLGNSGSHVLPQIFEQIFDCTL 918
Query 73 QMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEHPSLAE 132
M+K DF S+P+HRE FY +L+ +HCF GL LPS +L+A+V SL+WAFKHEHPS+AE
Sbjct 919 DMVKMDFHSFPEHREYFYEMLQKCTKHCFDGLLLLPSERLRAYVMSLIWAFKHEHPSVAE 978
Query 133 EGLQVTHEFLQRLI------------EGKREVLNDFCCNYYFSLMKEILLVL 172
GL V EFL L+ +G VL+ FC NYY+ L+KEIL VL
Sbjct 979 RGLTVVREFLNNLMLLDRRNDQSPGGQGMSAVLS-FCRNYYYLLLKEILGVL 1029
> pfa:PFC0135c exportin 1, putative; K14290 exportin-1
Length=1254
Score = 156 bits (394), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 68/161 (42%), Positives = 108/161 (67%), Gaps = 0/161 (0%)
Query 12 QLVTDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVIFDYIFDCT 71
Q+ +++L +L+ IL DYR++ P +D EV +L+S + +++ +LP + +Y+ T
Sbjct 948 QMTSNILNVLLETILVDYRDSNPHIKDAEVFSLLSTVFKKIENVTCPILPTVLNYVLLPT 1007
Query 72 LQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEHPSLA 131
+ MIK+DF SYP+HRE+FY L A +HCF LF+L S F++SL+WA KHEHPS+A
Sbjct 1008 IDMIKNDFSSYPEHREKFYNFLDACVRHCFDYLFTLDSEIFNTFIQSLLWAIKHEHPSVA 1067
Query 132 EEGLQVTHEFLQRLIEGKREVLNDFCCNYYFSLMKEILLVL 172
+ GL++T +FL +I K+E L +FC +Y+ ++ EIL L
Sbjct 1068 DHGLRITQQFLHNIIIKKKEYLEEFCKAFYYIILNEILKTL 1108
> tpv:TP02_0177 importin beta-related nuclear transport factor;
K14290 exportin-1
Length=1067
Score = 137 bits (346), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 61/136 (44%), Positives = 97/136 (71%), Gaps = 0/136 (0%)
Query 11 KQLVTDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVIFDYIFDC 70
+++ +L ++ ++ DYR++ +TRDYE+L+L + ++ RL ++ +SVL IF+Y+FD
Sbjct 925 NEMIVTLLDNLVVVMMGDYRDSVFETRDYEILSLSTKIIERLASNYSSVLVQIFNYVFDT 984
Query 71 TLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEHPSL 130
+L M+K DF +YPDHRE FY +L A + F + LP+ +L+ FV SLVWAFKHEHPS+
Sbjct 985 SLDMVKMDFHAYPDHREYFYEMLHKATKCSFDSVLLLPNERLRDFVMSLVWAFKHEHPSI 1044
Query 131 AEEGLQVTHEFLQRLI 146
A+ GL +T EF++ +I
Sbjct 1045 ADRGLLITLEFMKNII 1060
> hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homolog,
yeast); K14290 exportin-1
Length=1071
Score = 134 bits (336), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 62/172 (36%), Positives = 105/172 (61%), Gaps = 2/172 (1%)
Query 2 GGSLSAAARKQLVT-DMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVL 60
G +S + Q+V + + +L +L DY+ N P R+ EVL+ +++++++L HIT+ +
Sbjct 760 SGWVSRSNDPQMVAENFVPPLLDAVLIDYQRNVPAAREPEVLSTMAIIVNKLGGHITAEI 819
Query 61 PVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLV 120
P IFD +F+CTL MI DF+ YP+HR F+ LL+A N HCF ++P TQ K ++S++
Sbjct 820 PQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQAVNSHCFPAFLAIPPTQFKLVLDSII 879
Query 121 WAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYFSLMKEILLVL 172
WAFKH ++A+ GLQ+ LQ + + + F Y+ +++ I V+
Sbjct 880 WAFKHTMRNVADTGLQILFTLLQNVAQ-EEAAAQSFYQTYFCDILQHIFSVV 930
> xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog);
K14290 exportin-1
Length=1071
Score = 132 bits (331), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 57/147 (38%), Positives = 95/147 (64%), Gaps = 1/147 (0%)
Query 2 GGSLSAAARKQLVT-DMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVL 60
G +S ++ Q+V + + +L +L DY+ N P R+ EVL+ ++ ++++L HIT+ +
Sbjct 760 SGWVSRSSDPQMVAENFVPPLLDAVLIDYQRNVPAAREPEVLSTMATIVNKLGVHITAEI 819
Query 61 PVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLV 120
P IFD +F+CTL MI DF+ YP+HR F+ LL+A N HCF ++P Q K ++S++
Sbjct 820 PQIFDAVFECTLNMINKDFEEYPEHRTHFFLLLQAVNSHCFPAFLAIPPAQFKLVLDSII 879
Query 121 WAFKHEHPSLAEEGLQVTHEFLQRLIE 147
WAFKH ++A+ GLQ+ + LQ + +
Sbjct 880 WAFKHTMRNVADTGLQILYTLLQNVAQ 906
> mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolog
(yeast); K14290 exportin-1
Length=1071
Score = 132 bits (331), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 61/172 (35%), Positives = 104/172 (60%), Gaps = 2/172 (1%)
Query 2 GGSLSAAARKQLVT-DMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVL 60
G +S + Q+V + + +L +L DY+ N P R+ EVL+ +++++++L HIT+ +
Sbjct 760 SGWVSRSNDPQMVAENFVPPLLDAVLIDYQRNVPAAREPEVLSTMAIIVNKLGGHITAEI 819
Query 61 PVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLV 120
P IFD +F+CTL MI DF+ YP+HR F+ LL+A N HCF ++P Q K ++S++
Sbjct 820 PQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQAVNSHCFPAFLAIPPAQFKLVLDSII 879
Query 121 WAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYFSLMKEILLVL 172
WAFKH ++A+ GLQ+ LQ + + + F Y+ +++ I V+
Sbjct 880 WAFKHTMRNVADTGLQILFTLLQNVAQ-EEAAAQSFYQTYFCDILQHIFSVV 930
> dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b;
K14290 exportin-1
Length=1071
Score = 130 bits (328), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 61/172 (35%), Positives = 103/172 (59%), Gaps = 2/172 (1%)
Query 2 GGSLSAAARKQLV-TDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVL 60
G +S + Q+V + + +L +L DY+ N P R+ EVL+ ++ ++++L HIT +
Sbjct 760 SGWVSRSNDPQMVGENFVPPLLDAVLIDYQRNVPAAREPEVLSTMATIVNKLGGHITGEI 819
Query 61 PVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLV 120
P IFD +F+CTL MI DF+ YP+HR F+ LL+A N HCF ++P Q K ++S++
Sbjct 820 PQIFDAVFECTLNMINKDFEEYPEHRTHFFYLLQAVNSHCFPAFLAIPPAQFKLVLDSII 879
Query 121 WAFKHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYFSLMKEILLVL 172
WAFKH ++A+ GLQ+ + LQ + + + F Y+ +++ I V+
Sbjct 880 WAFKHTMRNVADTGLQILYTLLQNVAQ-EEAAAQSFYQTYFCDILQHIFSVV 930
> ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14290
exportin-1
Length=1076
Score = 122 bits (307), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 61/162 (37%), Positives = 96/162 (59%), Gaps = 6/162 (3%)
Query 11 KQLVTDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVIFDYIFDC 70
KQ V M+ QVL DY N PD R+ EVL+L + ++++ + +P+IF+ +F C
Sbjct 778 KQFVPPMMDQVLG----DYARNVPDARESEVLSLFATIINKYKVVMRDEVPLIFEAVFQC 833
Query 71 TLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEHPSL 130
TL+MI +F+ YP+HR +F++LL+A CF L L S QLK ++S++WAF+H ++
Sbjct 834 TLEMITKNFEDYPEHRLKFFSLLRAIATFCFRALIQLSSEQLKLVMDSVIWAFRHTERNI 893
Query 131 AEEGLQVTHEFLQRLIEGKREVLNDFCCNYYFSLMKEILLVL 172
AE GL + E L+ K + N F Y+ + +E+ VL
Sbjct 894 AETGLNLLLEMLKNF--QKSDFCNKFYQTYFLQIEQEVFAVL 933
> ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transporter/
receptor; K14290 exportin-1
Length=1075
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 60/162 (37%), Positives = 96/162 (59%), Gaps = 6/162 (3%)
Query 11 KQLVTDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVIFDYIFDC 70
KQ V M+ + +L DY N PD R+ EVL+L + ++++ + +P IF+ +F C
Sbjct 777 KQFVPPMM----ESVLGDYARNVPDARESEVLSLFATIINKYKATMLDDVPHIFEAVFQC 832
Query 71 TLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEHPSL 130
TL+MI +F+ YP+HR +F++LL+A CF L L S QLK ++S++WAF+H ++
Sbjct 833 TLEMITKNFEDYPEHRLKFFSLLRAIATFCFPALIKLSSPQLKLVMDSIIWAFRHTERNI 892
Query 131 AEEGLQVTHEFLQRLIEGKREVLNDFCCNYYFSLMKEILLVL 172
AE GL + E L+ + E N F +Y+ + +EI VL
Sbjct 893 AETGLNLLLEMLKNF--QQSEFCNQFYRSYFMQIEQEIFAVL 932
> sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1
Length=1084
Score = 119 bits (297), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 57/161 (35%), Positives = 89/161 (55%), Gaps = 1/161 (0%)
Query 13 LVTDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVIFDYIFDCTL 72
+V ++ +L +L+DY NN PD RD EVL ++ ++ ++ I + +I +F+CTL
Sbjct 783 VVKVLVEPLLNAVLEDYMNNVPDARDAEVLNCMTTVVEKVGHMIPQGVILILQSVFECTL 842
Query 73 QMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEHPSLAE 132
MI DF YP+HR FY LLK N+ F+ LP K FV+++ WAFKH + +
Sbjct 843 DMINKDFTEYPEHRVEFYKLLKVINEKSFAAFLELPPAAFKLFVDAICWAFKHNNRDVEV 902
Query 133 EGLQVTHEFLQRLIE-GKREVLNDFCCNYYFSLMKEILLVL 172
GLQ+ + ++ + G N+F NY+F + E VL
Sbjct 903 NGLQIALDLVKNIERMGNVPFANEFHKNYFFIFVSETFFVL 943
> cpv:cgd3_3060 exportin 1 ; K14290 exportin-1
Length=1266
Score = 117 bits (294), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 64/178 (35%), Positives = 100/178 (56%), Gaps = 20/178 (11%)
Query 15 TDMLGQVLQPI----LQDYRNNTPDTRDYEVLTLISVLMSRLDTHITS---VLPVIFDYI 67
T+ML ++ PI L+DY + ++ +VL L S ++ RL+ + + + I ++
Sbjct 914 TEMLQYIIHPIIIPVLEDYHACISEIKESQVLILSSTIIVRLNDIVKANNDLFNAIIYHL 973
Query 68 FDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAFKHEH 127
F+ TL MIK +F +YPDHRE FY+ L N+ CF LF+LP L ++ES++WA +HE
Sbjct 974 FESTLSMIKDNFHAYPDHREFFYSFLADCNEFCFLQLFNLPGNILTLYIESIIWAIRHEQ 1033
Query 128 PSLAEEGLQVTHEFLQRLIE-------------GKREVLNDFCCNYYFSLMKEILLVL 172
P++AE+GL V + FL LI + L FC +Y S+++EI VL
Sbjct 1034 PNMAEKGLIVLYNFLINLINHNSSKNNGIQSSCAQSNTLFQFCHAFYLSIIREIFCVL 1091
> cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family
member (xpo-1); K14290 exportin-1
Length=1080
Score = 109 bits (272), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 52/168 (30%), Positives = 95/168 (56%), Gaps = 1/168 (0%)
Query 5 LSAAARKQLVTD-MLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVI 63
+S +L+ D ++ + +L DY+ N P R+ +VL+L+S+L+++L + + +P I
Sbjct 773 ISKNGDAKLILDSIVPPLFDAVLFDYQKNVPQAREPKVLSLLSILVTQLGSLLCPQVPSI 832
Query 64 FDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFSGLFSLPSTQLKAFVESLVWAF 123
+F C++ MI D +++P+HR F+ L+ + Q CF +P L ++++VWAF
Sbjct 833 LSAVFQCSIDMINKDMEAFPEHRTNFFELVLSLVQECFPVFMEMPPEDLGTVIDAVVWAF 892
Query 124 KHEHPSLAEEGLQVTHEFLQRLIEGKREVLNDFCCNYYFSLMKEILLV 171
+H ++AE GL + E L R+ E ++ F YY L+K +L V
Sbjct 893 QHTMRNVAEIGLDILKELLARVSEQDDKIAQPFYKRYYIDLLKHVLAV 940
> hsa:8450 CUL4B, DKFZp686F1470, KIAA0695, MRXHF2, MRXS15, MRXSC,
SFM2; cullin 4B; K10609 cullin 4
Length=895
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 61/143 (42%), Gaps = 24/143 (16%)
Query 4 SLSAAARKQLVTDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVI 63
SL A KQL LG+ L ILQ NN D + L+L+ L SR+ + +L
Sbjct 421 SLIATVEKQL----LGEHLTAILQKGLNNLLDENRIQDLSLLYQLFSRVRGGVQVLLQQW 476
Query 64 FDYI--FDCTL--------QMIKS--DFQSYPDH--------RERFYALLKAANQHCFSG 103
+YI F T+ M++ DF+ DH E+F +K A + +
Sbjct 477 IEYIKAFGSTIVINPEKDKTMVQELLDFKDKVDHIIDICFLKNEKFINAMKEAFETFINK 536
Query 104 LFSLPSTQLKAFVESLVWAFKHE 126
+ P+ + +V+S + A E
Sbjct 537 RPNKPAELIAKYVDSKLRAGNKE 559
> dre:560313 cul4b, im:7140950, im:7150765; cullin 4B; K10609
cullin 4
Length=864
Score = 30.0 bits (66), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 61/143 (42%), Gaps = 24/143 (16%)
Query 4 SLSAAARKQLVTDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVI 63
SL A KQL LG+ L ILQ NN D + L+L+ L SR+ + +L
Sbjct 390 SLIATVEKQL----LGEHLTAILQKGLNNLLDENRIQDLSLLYQLFSRVRGGVQVLLQHW 445
Query 64 FDYI--FDCTL--------QMIKS--DFQSYPDH--------RERFYALLKAANQHCFSG 103
+YI F T+ M++ DF+ DH E+F +K A + +
Sbjct 446 IEYIKAFGSTIVINPEKDKTMVQELLDFKDKVDHIIDVCFMKNEKFVNGMKEAFETFINK 505
Query 104 LFSLPSTQLKAFVESLVWAFKHE 126
+ P+ + +V+S + A E
Sbjct 506 RPNKPAELIAKYVDSKLRAGNKE 528
> cel:Y4C6B.5 hypothetical protein; K14613 MFS transporter, PCFT/HCP
family, solute carrier family 46 (folate transporter),
member 1/3
Length=469
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 37/77 (48%), Gaps = 2/77 (2%)
Query 43 TLISVLMSRLDTHITSV-LPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCF 101
+L++ L+ T S+ +P+I YI C L I FQSY H ++ L+ A
Sbjct 79 SLVTTLLLGAATDYWSIKIPLIIPYI-GCILGTINYVFQSYFIHTSVYFLLISDALFGLC 137
Query 102 SGLFSLPSTQLKAFVES 118
G ++ ST L V++
Sbjct 138 GGFIAIISTTLTYGVKT 154
> ath:AT1G05680 UDP-glucoronosyl/UDP-glucosyl transferase family
protein
Length=453
Score = 29.3 bits (64), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 34/77 (44%), Gaps = 14/77 (18%)
Query 49 MSRLDTHITSVLPVIFD-----------YIFDCTLQMIKSDFQSYPDHRERFYA---LLK 94
M R++T I + LP + + ++D T+ + SY F+ L+
Sbjct 78 MERVETSIKNTLPKLVEDMKLSGNPPRAIVYDSTMPWLLDVAHSYGLSGAVFFTQPWLVT 137
Query 95 AANQHCFSGLFSLPSTQ 111
A H F G FS+PST+
Sbjct 138 AIYYHVFKGSFSVPSTK 154
> mmu:72584 Cul4b, 2700050M05Rik, AA409770, KIAA0695, mKIAA0695;
cullin 4B; K10609 cullin 4
Length=970
Score = 28.9 bits (63), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 62/143 (43%), Gaps = 24/143 (16%)
Query 4 SLSAAARKQLVTDMLGQVLQPILQDYRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVI 63
SL A+ KQL LG+ L ILQ N+ D + L+L+ L SR+ + +L
Sbjct 496 SLIASVEKQL----LGEHLTAILQKGLNSLLDENRIQDLSLLYQLFSRVRGGVQVLLQQW 551
Query 64 FDYI--FDCTL--------QMIKS--DFQSYPDH--------RERFYALLKAANQHCFSG 103
+YI F T+ M++ DF+ DH E+F +K A + +
Sbjct 552 IEYIKAFGSTIVINPEKDKTMVQELLDFKDKVDHIIDTCFLKNEKFINAMKEAFETFINK 611
Query 104 LFSLPSTQLKAFVESLVWAFKHE 126
+ P+ + +V+S + A E
Sbjct 612 RPNKPAELIAKYVDSKLRAGNKE 634
> cel:K04F1.4 srw-91; Serpentine Receptor, class W family member
(srw-91)
Length=350
Score = 28.9 bits (63), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 29 YRNNTPDTRDYEVLTLISVLMSRLDTHITSVLPVIFD 65
Y+NNT T + ++ S+ ++ L T I +V VI+
Sbjct 265 YKNNTEKTTNLVIIMTFSIFLASLPTGIATVFQVIYS 301
> ath:AT1G27595 hypothetical protein; K06100 symplekin
Length=961
Score = 28.9 bits (63), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 22/96 (22%), Positives = 35/96 (36%), Gaps = 4/96 (4%)
Query 43 TLISVLMSRLDTHITSVLPVIFDYIFDCTLQMIKSDFQSYPDHRERFYALLKAANQHCFS 102
T + +L R P + D++ + ++++ P F + H F
Sbjct 830 TPLPLLFMRTVIQAIDAFPTLVDFVMEILSKLVRKQIWRLPKLWPGFLKCVSQTKPHSFP 889
Query 103 GLFSLPSTQLKAFVESLVWAFKHEHPSLAEEGLQVT 138
L LP QL ES++ F PSL Q T
Sbjct 890 VLLELPVPQL----ESIMKKFPDLRPSLTAYANQPT 921
Lambda K H
0.325 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40