bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7502_orf2
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
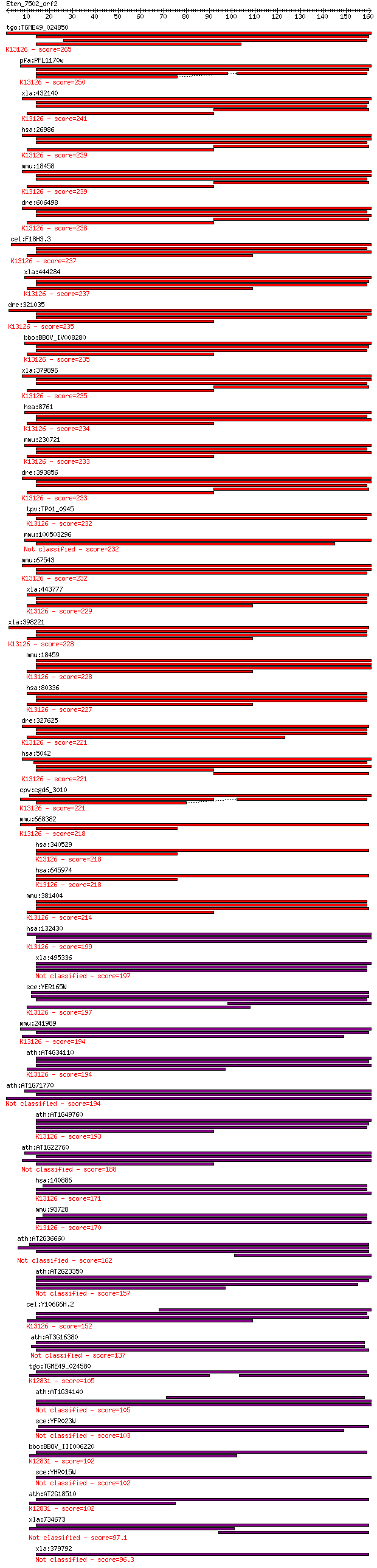
tgo:TGME49_024850 polyadenylate-binding protein, putative ; K1... 265 3e-71
pfa:PFL1170w polyadenylate-binding protein, putative; K13126 p... 250 2e-66
xla:432140 pabpc1-b, MGC130736, MGC79060, pab1, pabp, pabp1, p... 241 7e-64
hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A) b... 239 2e-63
mmu:18458 Pabpc1, PABP, Pabp1, PabpI, Pabpl1, ePAB; poly(A) bi... 239 2e-63
dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09... 238 8e-63
cel:F18H3.3 pab-2; PolyA Binding protein family member (pab-2)... 237 1e-62
xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) bindin... 237 1e-62
dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cyt... 235 4e-62
bbo:BBOV_IV008280 23.m06515; polyadenylate binding protein; K1... 235 6e-62
xla:379896 pabpc1-a, MGC53109, pab1, pabp, pabp1, pabpc1, pabp... 235 6e-62
hsa:8761 PABPC4, APP-1, APP1, FLJ43938, PABP4, iPABP; poly(A) ... 234 1e-61
mmu:230721 Pabpc4, MGC11665, MGC6685; poly(A) binding protein,... 233 2e-61
dre:393856 pabpc1b, MGC77608, pabpc1, zgc:77608; poly A bindin... 233 2e-61
tpv:TP01_0945 polyadenylate binding protein; K13126 polyadenyl... 232 5e-61
mmu:100503296 polyadenylate-binding protein 4-like 232 5e-61
mmu:67543 Pabpc6, 4932702K14Rik, AI428050, MGC132900, Pabpc3; ... 232 5e-61
xla:443777 pabpc1l-b, MGC81363, epab, epabp-b, pabpc1l1; poly(... 229 3e-60
xla:398221 pabpc1l-a, MGC82273, ePAB, epabp-a, pabpc1l1; poly(... 228 7e-60
mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,... 228 9e-60
hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053, PA... 227 1e-59
dre:327625 pabpc1l, fi20g03, pab-2, wu:fi20g03, zgc:55855; pol... 221 7e-58
hsa:5042 PABPC3, PABP3, PABPL3, tPABP; poly(A) binding protein... 221 1e-57
cpv:cgd6_3010 poly(a)-binding protein fabm ; K13126 polyadenyl... 221 1e-57
mmu:668382 Pabpc1l2b-ps, EG668382, Pabpc1l2b, Rbm32b; poly(A) ... 218 6e-57
hsa:340529 PABPC1L2A, MGC168104, PABPC1L2B, RBM32A; poly(A) bi... 218 7e-57
hsa:645974 PABPC1L2B, MGC168104, PABPC1L2A, RBM32B; poly(A) bi... 218 7e-57
mmu:381404 Pabpc1l, 1810053B01Rik, Epab; poly(A) binding prote... 214 8e-56
hsa:132430 PABPC4L, DKFZp686J06116; poly(A) binding protein, c... 199 3e-51
xla:495336 hypothetical LOC495336 197 1e-50
sce:YER165W PAB1; Pab1p; K13126 polyadenylate-binding protein 197 1e-50
mmu:241989 Pabpc4l, C330050A14Rik, EG241989; poly(A) binding p... 194 7e-50
ath:AT4G34110 PAB2; PAB2 (POLY(A) BINDING 2); RNA binding / tr... 194 8e-50
ath:AT1G71770 PAB5; PAB5 (POLY(A)-BINDING PROTEIN 5); RNA bind... 194 1e-49
ath:AT1G49760 PAB8; PAB8 (POLY(A) BINDING PROTEIN 8); RNA bind... 193 2e-49
ath:AT1G22760 PAB3; PAB3 (POLY(A) BINDING PROTEIN 3); RNA bind... 188 7e-48
hsa:140886 PABPC5, FLJ51677, PABP5; poly(A) binding protein, c... 171 1e-42
mmu:93728 Pabpc5, C820015E17, MGC130340, MGC130341; poly(A) bi... 170 2e-42
ath:AT2G36660 PAB7; PAB7 (POLY(A) BINDING PROTEIN 7); RNA bind... 162 6e-40
ath:AT2G23350 PAB4; PAB4 (POLY(A) BINDING PROTEIN 4); RNA bind... 157 2e-38
cel:Y106G6H.2 pab-1; PolyA Binding protein family member (pab-... 152 3e-37
ath:AT3G16380 PAB6; PAB6 (POLY(A) BINDING PROTEIN 6); RNA bind... 137 1e-32
tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12... 105 8e-23
ath:AT1G34140 PAB1; PAB1 (POLY(A) BINDING PROTEIN 1); RNA bind... 105 9e-23
sce:YFR023W PES4; Pes4p 103 2e-22
bbo:BBOV_III006220 17.m07552; RNA recognition motif domaining ... 102 5e-22
sce:YHR015W MIP6; Mip6p 102 6e-22
ath:AT2G18510 emb2444 (embryo defective 2444); RNA binding / n... 102 7e-22
xla:734673 hypothetical protein MGC116464 97.1 2e-20
xla:379792 sf3b4, MGC52742, spx; splicing factor 3b, subunit 4... 96.3 4e-20
> tgo:TGME49_024850 polyadenylate-binding protein, putative ;
K13126 polyadenylate-binding protein
Length=768
Score = 265 bits (678), Expect = 3e-71, Method: Compositional matrix adjust.
Identities = 123/160 (76%), Positives = 141/160 (88%), Gaps = 3/160 (1%)
Query 1 DAGAGPHPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVN 60
+AGA +VSPSLY+GDLH +VTEAMLFEVFN+VGPVTSIRVCRD+V+RRSLGYAYVN
Sbjct 74 NAGAA---NFVSPSLYVGDLHQDVTEAMLFEVFNSVGPVTSIRVCRDTVTRRSLGYAYVN 130
Query 61 YQSLGDAERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDT 120
YQ + DAER+LD LN+ +I+GQPCRIMW RDPSLR+SG GN+FVKNLDK+IDNKALYDT
Sbjct 131 YQGIQDAERSLDTLNYTVIKGQPCRIMWCHRDPSLRKSGNGNIFVKNLDKNIDNKALYDT 190
Query 121 FSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
FSLFGNILSCKV VD+ S+GYGFVHYE E SARSAI+K
Sbjct 191 FSLFGNILSCKVAVDDNGHSKGYGFVHYENEESARSAIDK 230
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 43/151 (28%), Positives = 83/151 (54%), Gaps = 9/151 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++++ +L + L++ F+ G + S +V D + S GY +V+Y++ A A+D
Sbjct 172 NIFVKNLDKNIDNKALYDTFSLFGNILSCKVAVDD-NGHSKGYGFVHYENEESARSAIDK 230
Query 74 LNFALIRGQPCR----IMWSQRDPSLRRSGAGNVFVKNLDKSIDNKA-LYDTFSLFGNIL 128
+N LI G+ I ++RD +L + NV++KN+ + ++++ L +TFS FG+I
Sbjct 231 VNGMLIGGKTVYVGPFIRRAERD-NLAEAKYTNVYIKNMPSAWEDESRLRETFSKFGSIT 289
Query 129 SCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
S V D + R + F ++ SA++A+E
Sbjct 290 SLVVRKDP--KGRLFAFCNFADHDSAKAAVE 318
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 48/174 (27%), Positives = 76/174 (43%), Gaps = 44/174 (25%)
Query 26 EAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDALNF-------AL 78
E+ L E F+ G +TS+ V +D R +A+ N+ A+ A++ALN A+
Sbjct 275 ESRLRETFSKFGSITSLVVRKDPKGRL---FAFCNFADHDSAKAAVEALNGKRVTDAGAI 331
Query 79 IRGQPC---------------RIMW-------SQRDPSLR------------RSGAGNVF 104
G+ +I++ + R LR R N++
Sbjct 332 KEGEDSGAEEKEEEGQKREGDQILFVGPHQSKAHRSAMLRAKFEQMNQDRNDRFQGVNLY 391
Query 105 VKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
+KN+D SID++ L F FG+I S KV DE SR +GFV + + A A+
Sbjct 392 IKNMDDSIDDEKLRQLFEPFGSITSAKVMRDERGVSRCFGFVCFMSPEEATKAV 445
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 55/108 (50%), Gaps = 21/108 (19%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDS--VSRRSLGYAYVNYQSLGDAERAL 71
+LYI ++ + + L ++F G +TS +V RD VSR + +V + S +A +A+
Sbjct 389 NLYIKNMDDSIDDEKLRQLFEPFGSITSAKVMRDERGVSR---CFGFVCFMSPEEATKAV 445
Query 72 DALNFALIRGQPC----------RIMWSQ---RDPSLRRSGA---GNV 103
++ L++G+P R+M Q R PSLR + A GNV
Sbjct 446 TEMHLKLVKGKPLYVGLAERREQRLMRLQQRFRLPSLRPAAAALPGNV 493
> pfa:PFL1170w polyadenylate-binding protein, putative; K13126
polyadenylate-binding protein
Length=875
Score = 250 bits (638), Expect = 2e-66, Method: Composition-based stats.
Identities = 111/154 (72%), Positives = 134/154 (87%), Gaps = 0/154 (0%)
Query 7 HPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGD 66
HP++ + SLY+GDL+ +VTEA+L+E+FN VG V+SIRVCRDSV+R+SLGYAYVNY +L D
Sbjct 10 HPSFSTASLYVGDLNEDVTEAVLYEIFNTVGHVSSIRVCRDSVTRKSLGYAYVNYHNLAD 69
Query 67 AERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGN 126
AERALD LN+ I+GQP R+MWS RDPSLR+SG GN+FVKNLDKSIDNKAL+DTFS+FGN
Sbjct 70 AERALDTLNYTNIKGQPARLMWSHRDPSLRKSGTGNIFVKNLDKSIDNKALFDTFSMFGN 129
Query 127 ILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
ILSCKV DEF +S+ YGFVHYE E SA+ AIEK
Sbjct 130 ILSCKVATDEFGKSKSYGFVHYEDEESAKEAIEK 163
Score = 53.1 bits (126), Expect = 4e-07, Method: Composition-based stats.
Identities = 34/148 (22%), Positives = 74/148 (50%), Gaps = 5/148 (3%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++++ +L + LF+ F+ G + S +V D + S Y +V+Y+ A+ A++
Sbjct 105 NIFVKNLDKSIDNKALFDTFSMFGNILSCKVATDEFGK-SKSYGFVHYEDEESAKEAIEK 163
Query 74 LNFALIRGQPCRI--MWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCK 131
+N + + + + + + + N++VKN S+ L F+ +G I S
Sbjct 164 VNGVQLGSKNVYVGPFIKKSERATNDTKFTNLYVKNFPDSVTETHLRQLFNPYGEITSMI 223
Query 132 VGVDEFNRSRGYGFVHYETEASARSAIE 159
V +D N++R + F++Y SA++A++
Sbjct 224 VKMD--NKNRKFCFINYADAESAKNAMD 249
Score = 51.6 bits (122), Expect = 1e-06, Method: Composition-based stats.
Identities = 24/57 (42%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 102 NVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
N+++KNLD ID+ L + F FG I S KV DE +S+G+GFV + ++ A A+
Sbjct 451 NLYIKNLDDGIDDIMLRELFEPFGTITSAKVMRDEKEQSKGFGFVCFASQEEANKAV 507
Score = 46.2 bits (108), Expect = 4e-05, Method: Composition-based stats.
Identities = 26/85 (30%), Positives = 48/85 (56%), Gaps = 2/85 (2%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LYI +L + + ML E+F G +TS +V RD +S G+ +V + S +A +A+
Sbjct 451 NLYIKNLDDGIDDIMLRELFEPFGTITSAKVMRDE-KEQSKGFGFVCFASQEEANKAVTE 509
Query 74 LNFALIRGQPCRI-MWSQRDPSLRR 97
++ +I G+P + + +R+ L R
Sbjct 510 MHLKIINGKPLYVGLAEKREQRLSR 534
Score = 40.4 bits (93), Expect = 0.003, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 3/62 (4%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+ + VTE L ++FN G +TS+ V D+ +R+ + ++NY A+ A+D
Sbjct 194 NLYVKNFPDSVTETHLRQLFNPYGEITSMIVKMDNKNRK---FCFINYADAESAKNAMDN 250
Query 74 LN 75
LN
Sbjct 251 LN 252
> xla:432140 pabpc1-b, MGC130736, MGC79060, pab1, pabp, pabp1,
pabpc2; poly(A) binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=633
Score = 241 bits (615), Expect = 7e-64, Method: Compositional matrix adjust.
Identities = 109/153 (71%), Positives = 130/153 (84%), Gaps = 1/153 (0%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
P+Y SLY+GDLHP+VTEAML+E F+ GP+ SIRVCRD ++RRSLGYAYVN+Q DA
Sbjct 6 PSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADA 65
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
ERALD +NF +I+G+P RIMWSQRDPSLR+SG GN+F+KNLDKSIDNKALYDTFS FGNI
Sbjct 66 ERALDTMNFDVIKGKPVRIMWSQRDPSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNI 125
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LSCKV DE N S+GYGFVH+ET+ +A AI+K
Sbjct 126 LSCKVVCDE-NGSKGYGFVHFETQEAAERAIDK 157
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 45/152 (29%), Positives = 82/152 (53%), Gaps = 8/152 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GY +V++++ AERA+D
Sbjct 100 NIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENG--SKGYGFVHFETQEAAERAIDK 157
Query 74 LNFALIRGQPCRI----MWSQRDPSL--RRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + +R+ L R NV++KN + +D++ L + F +G
Sbjct 158 MNGMLLNDRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDDERLKEWFGQYGAA 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
LS KV D+ +SRG+GFV +E A+ A++
Sbjct 218 LSVKVMTDDHGKSRGFGFVSFERHEDAQKAVD 249
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 47/161 (29%), Positives = 82/161 (50%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L E F G S++V D + S G+ +V+++ DA++A+D
Sbjct 192 NVYIKNFGEDMDDERLKEWFGQYGAALSVKVMTDDHGK-SRGFGFVSFERHEDAQKAVDD 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q R L+R N++VKNLD ID++ L
Sbjct 251 MNGKDLNGKAIFVGRAQKKVERQTELKRKFEQMKQDRITRYQGVNLYVKNLDDGIDDERL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
F+ FG+I S KV + E RS+G+GFV + + A A+
Sbjct 311 RKEFTPFGSITSAKVMM-EGGRSKGFGFVCFSSPEEATKAV 350
Score = 37.7 bits (86), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 1/69 (1%)
Query 92 DPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFN-RSRGYGFVHYET 150
+PS +++V +L + LY+ FS G ILS +V D RS GY +V+++
Sbjct 2 NPSAPSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQ 61
Query 151 EASARSAIE 159
A A A++
Sbjct 62 PADAERALD 70
Score = 35.4 bits (80), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 42/82 (51%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F G +TS +V + RS G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDGIDDERLRKEFTPFGSITSAKVMME--GGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 349 AVTEMNGRIVATKPLYVALAQR 370
> hsa:26986 PABPC1, PAB1, PABP, PABP1, PABPC2, PABPL1; poly(A)
binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=636
Score = 239 bits (611), Expect = 2e-63, Method: Composition-based stats.
Identities = 110/153 (71%), Positives = 130/153 (84%), Gaps = 1/153 (0%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
P+Y SLY+GDLHP+VTEAML+E F+ GP+ SIRVCRD ++RRSLGYAYVN+Q DA
Sbjct 6 PSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADA 65
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
ERALD +NF +I+G+P RIMWSQRDPSLR+SG GN+F+KNLDKSIDNKALYDTFS FGNI
Sbjct 66 ERALDTMNFDVIKGKPVRIMWSQRDPSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNI 125
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LSCKV DE N S+GYGFVH+ET+ +A AIEK
Sbjct 126 LSCKVVCDE-NGSKGYGFVHFETQEAAERAIEK 157
Score = 76.6 bits (187), Expect = 4e-14, Method: Composition-based stats.
Identities = 46/153 (30%), Positives = 83/153 (54%), Gaps = 8/153 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GY +V++++ AERA++
Sbjct 100 NIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDE--NGSKGYGFVHFETQEAAERAIEK 157
Query 74 LNFALIRGQPCRI----MWSQRDPSL--RRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + +R+ L R NV++KN + +D++ L D F FG
Sbjct 158 MNGMLLNDRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDDERLKDLFGKFGPA 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LS KV DE +S+G+GFV +E A+ A+++
Sbjct 218 LSVKVMTDESGKSKGFGFVSFERHEDAQKAVDE 250
Score = 72.8 bits (177), Expect = 5e-13, Method: Composition-based stats.
Identities = 49/161 (30%), Positives = 84/161 (52%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L ++F GP S++V D S +S G+ +V+++ DA++A+D
Sbjct 192 NVYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDE-SGKSKGFGFVSFERHEDAQKAVDE 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q R L+R N++VKNLD ID++ L
Sbjct 251 MNGKELNGKQIYVGRAQKKVERQTELKRKFEQMKQDRITRYQGVNLYVKNLDDGIDDERL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
FS FG I S KV + E RS+G+GFV + + A A+
Sbjct 311 RKEFSPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAV 350
Score = 37.0 bits (84), Expect = 0.026, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 1/69 (1%)
Query 92 DPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFN-RSRGYGFVHYET 150
+PS +++V +L + LY+ FS G ILS +V D RS GY +V+++
Sbjct 2 NPSAPSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQ 61
Query 151 EASARSAIE 159
A A A++
Sbjct 62 PADAERALD 70
Score = 35.8 bits (81), Expect = 0.068, Method: Composition-based stats.
Identities = 21/82 (25%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F+ G +TS +V + RS G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDGIDDERLRKEFSPFGTITSAKVMME--GGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 349 AVTEMNGRIVATKPLYVALAQR 370
> mmu:18458 Pabpc1, PABP, Pabp1, PabpI, Pabpl1, ePAB; poly(A)
binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=636
Score = 239 bits (611), Expect = 2e-63, Method: Composition-based stats.
Identities = 110/153 (71%), Positives = 130/153 (84%), Gaps = 1/153 (0%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
P+Y SLY+GDLHP+VTEAML+E F+ GP+ SIRVCRD ++RRSLGYAYVN+Q DA
Sbjct 6 PSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADA 65
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
ERALD +NF +I+G+P RIMWSQRDPSLR+SG GN+F+KNLDKSIDNKALYDTFS FGNI
Sbjct 66 ERALDTMNFDVIKGKPVRIMWSQRDPSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNI 125
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LSCKV DE N S+GYGFVH+ET+ +A AIEK
Sbjct 126 LSCKVVCDE-NGSKGYGFVHFETQEAAERAIEK 157
Score = 74.7 bits (182), Expect = 1e-13, Method: Composition-based stats.
Identities = 45/153 (29%), Positives = 83/153 (54%), Gaps = 8/153 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GY +V++++ AERA++
Sbjct 100 NIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDE--NGSKGYGFVHFETQEAAERAIEK 157
Query 74 LNFALIRGQPCRI----MWSQRDPSL--RRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + +R+ L R NV++KN + +D++ L + F FG
Sbjct 158 MNGMLLNDRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDDERLKELFGKFGPA 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LS KV DE +S+G+GFV +E A+ A+++
Sbjct 218 LSVKVMTDESGKSKGFGFVSFERHEDAQKAVDE 250
Score = 73.9 bits (180), Expect = 2e-13, Method: Composition-based stats.
Identities = 50/161 (31%), Positives = 84/161 (52%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L E+F GP S++V D S +S G+ +V+++ DA++A+D
Sbjct 192 NVYIKNFGEDMDDERLKELFGKFGPALSVKVMTDE-SGKSKGFGFVSFERHEDAQKAVDE 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q R L+R N++VKNLD ID++ L
Sbjct 251 MNGKELNGKQIYVGRAQKKVERQTELKRKFEQMKQDRITRYQGVNLYVKNLDDGIDDERL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
FS FG I S KV + E RS+G+GFV + + A A+
Sbjct 311 RKEFSPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAV 350
Score = 37.0 bits (84), Expect = 0.026, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 1/69 (1%)
Query 92 DPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFN-RSRGYGFVHYET 150
+PS +++V +L + LY+ FS G ILS +V D RS GY +V+++
Sbjct 2 NPSAPSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQ 61
Query 151 EASARSAIE 159
A A A++
Sbjct 62 PADAERALD 70
Score = 35.8 bits (81), Expect = 0.069, Method: Composition-based stats.
Identities = 21/82 (25%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F+ G +TS +V + RS G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDGIDDERLRKEFSPFGTITSAKVMME--GGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 349 AVTEMNGRIVATKPLYVALAQR 370
> dre:606498 pabpc1a, pabpc1, wu:fb16a02, wu:fi19b08, wu:fj12d09,
wu:fj61f06, zgc:109879; poly A binding protein, cytoplasmic
1 a; K13126 polyadenylate-binding protein
Length=634
Score = 238 bits (606), Expect = 8e-63, Method: Composition-based stats.
Identities = 110/153 (71%), Positives = 129/153 (84%), Gaps = 1/153 (0%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
P+Y SLY+GDLHP+VTEAML+E F+ GP+ SIRVCRD ++RRSLGYAYVN+Q DA
Sbjct 6 PSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMMTRRSLGYAYVNFQQPADA 65
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
ERALD +NF +I+G+P RIMWSQRDPSLR+SG GN+F+KNLDKSIDNKALYDTFS FGNI
Sbjct 66 ERALDTMNFDVIKGRPVRIMWSQRDPSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNI 125
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LSCKV DE N S+GYGFVH+ET +A AIEK
Sbjct 126 LSCKVVCDE-NGSKGYGFVHFETHEAAERAIEK 157
Score = 75.9 bits (185), Expect = 5e-14, Method: Composition-based stats.
Identities = 52/161 (32%), Positives = 84/161 (52%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L E+F GP SIRV D S +S G+ +V+++ DA+RA+D
Sbjct 192 NVYIKNFGEDMDDEKLKEIFCKYGPALSIRVMTDD-SGKSKGFGFVSFERHEDAQRAVDE 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q R L+R N++VKNLD +D++ L
Sbjct 251 MNGKEMNGKQVYVGRAQKKGERQTELKRKFEQMKQDRMTRYQGVNLYVKNLDDGLDDERL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
FS FG I S KV + E RS+G+GFV + + A A+
Sbjct 311 RKEFSPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAV 350
Score = 70.1 bits (170), Expect = 3e-12, Method: Composition-based stats.
Identities = 42/153 (27%), Positives = 82/153 (53%), Gaps = 8/153 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GY +V++++ AERA++
Sbjct 100 NIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDE--NGSKGYGFVHFETHEAAERAIEK 157
Query 74 LNFALIRGQPCRI-MWSQRDPSLRRSGA-----GNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + + R GA NV++KN + +D++ L + F +G
Sbjct 158 MNGMLLNDRKVFVGRFKSRKEREAEMGARAKEFTNVYIKNFGEDMDDEKLKEIFCKYGPA 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LS +V D+ +S+G+GFV +E A+ A+++
Sbjct 218 LSIRVMTDDSGKSKGFGFVSFERHEDAQRAVDE 250
Score = 37.0 bits (84), Expect = 0.025, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 1/69 (1%)
Query 92 DPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFN-RSRGYGFVHYET 150
+PS +++V +L + LY+ FS G ILS +V D RS GY +V+++
Sbjct 2 NPSAPSYPMASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMMTRRSLGYAYVNFQQ 61
Query 151 EASARSAIE 159
A A A++
Sbjct 62 PADAERALD 70
Score = 35.0 bits (79), Expect = 0.11, Method: Composition-based stats.
Identities = 21/82 (25%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F+ G +TS +V + RS G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDGLDDERLRKEFSPFGTITSAKVMME--GGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 349 AVTEMNGRIVATKPLYVALAQR 370
> cel:F18H3.3 pab-2; PolyA Binding protein family member (pab-2);
K13126 polyadenylate-binding protein
Length=566
Score = 237 bits (604), Expect = 1e-62, Method: Composition-based stats.
Identities = 109/158 (68%), Positives = 130/158 (82%), Gaps = 0/158 (0%)
Query 3 GAGPHPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQ 62
G P+ TY SLYIGDLHP+V+EAMLFE F+ GPV SIRVCRD+ SR SLGYAYVN+Q
Sbjct 47 GYPPNATYSMASLYIGDLHPDVSEAMLFEKFSMAGPVLSIRVCRDNTSRLSLGYAYVNFQ 106
Query 63 SLGDAERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFS 122
DAERALD +NF +I G+P RIMWSQRDP+ RR+G GN+F+KNLD+ IDNK++YDTFS
Sbjct 107 QPADAERALDTMNFEVIHGRPMRIMWSQRDPAARRAGNGNIFIKNLDRVIDNKSVYDTFS 166
Query 123 LFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LFGNILSCKV D+ S+GYGFVH+ETE SA++AIEK
Sbjct 167 LFGNILSCKVATDDEGNSKGYGFVHFETEHSAQTAIEK 204
Score = 70.5 bits (171), Expect = 2e-12, Method: Composition-based stats.
Identities = 45/163 (27%), Positives = 83/163 (50%), Gaps = 19/163 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++++ + + + L +F+ G +TS V D+ + G+ +V + A +A++
Sbjct 239 NVFVKNFGEHLDQEKLSAMFSKFGEITSAVVMTDAQGKPK-GFGFVAFADQDAAGQAVEK 297
Query 74 LNFALIRGQPCRIMW------SQRDPSLRRSGAG------------NVFVKNLDKSIDNK 115
LN +++ G C++ S+R L+R N++VKN+++ +++
Sbjct 298 LNDSILEGTDCKLSVCRAQKKSERSAELKRKYEALKQERVQRYQGVNLYVKNIEEDLNDD 357
Query 116 ALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
L D FS FG I S KV VDE RS+G+GFV +E A +A+
Sbjct 358 GLRDHFSSFGTITSAKVMVDENGRSKGFGFVCFEKPEEATAAV 400
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 44/153 (28%), Positives = 78/153 (50%), Gaps = 7/153 (4%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + +++ F+ G + S +V D S GY +V++++ A+ A++
Sbjct 146 NIFIKNLDRVIDNKSVYDTFSLFGNILSCKVATDD-EGNSKGYGFVHFETEHSAQTAIEK 204
Query 74 LNFALIRGQPCRI----MWSQRDPSLRRSGAG--NVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + +QR L SG NVFVKN + +D + L FS FG I
Sbjct 205 VNGMLLSDKKVYVGKFQPRAQRMKELGESGLKYTNVFVKNFGEHLDQEKLSAMFSKFGEI 264
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
S V D + +G+GFV + + +A A+EK
Sbjct 265 TSAVVMTDAQGKPKGFGFVAFADQDAAGQAVEK 297
Score = 38.1 bits (87), Expect = 0.012, Method: Composition-based stats.
Identities = 24/99 (24%), Positives = 53/99 (53%), Gaps = 2/99 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ ++ ++ + L + F++ G +TS +V D + RS G+ +V ++ +A
Sbjct 340 YQGVNLYVKNIEEDLNDDGLRDHFSSFGTITSAKVMVDE-NGRSKGFGFVCFEKPEEATA 398
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNL 108
A+ +N +I +P + +QR RR+ + +++ L
Sbjct 399 AVTDMNSKMIGAKPLYVALAQRKED-RRAQLASQYMQRL 436
> xla:444284 pabpc4, MGC80927, PABP, ePAB, ePABP; poly(A) binding
protein, cytoplasmic 4 (inducible form); K13126 polyadenylate-binding
protein
Length=626
Score = 237 bits (604), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 110/152 (72%), Positives = 128/152 (84%), Gaps = 1/152 (0%)
Query 9 TYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAE 68
+Y SLY+GDLHP+VTEAML+E F+ GPV SIRVCRD ++RRSLGYAYVN+Q DAE
Sbjct 7 SYPMASLYVGDLHPDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAE 66
Query 69 RALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNIL 128
RALD +NF +I+G+P RIMWSQRDPSLR+SG GNVF+KNLDKSIDNKALYDTFS FGNIL
Sbjct 67 RALDTMNFDVIKGKPIRIMWSQRDPSLRKSGVGNVFIKNLDKSIDNKALYDTFSAFGNIL 126
Query 129 SCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
SCKV DE N S+GY FVH+ET+ +A AIEK
Sbjct 127 SCKVVCDE-NGSKGYAFVHFETQDAADRAIEK 157
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 46/155 (29%), Positives = 84/155 (54%), Gaps = 14/155 (9%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GYA+V++++ A+RA++
Sbjct 100 NVFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENG--SKGYAFVHFETQDAADRAIEK 157
Query 74 LNFALIRGQP-------CRIMWSQRDPSL--RRSGAGNVFVKNLDKSIDNKALYDTFSLF 124
+N L+ + CR +R+ L + NV++KN + +D++ L +TFS +
Sbjct 158 MNGMLLNDRKVFVGRFKCR---REREAELGAKAKEFTNVYIKNFGEDMDDERLKETFSKY 214
Query 125 GNILSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
G LS KV D +S+G+GFV +E A A++
Sbjct 215 GKTLSVKVMTDPSGKSKGFGFVSFERHEDANKAVD 249
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 48/161 (29%), Positives = 85/161 (52%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L E F+ G S++V D S +S G+ +V+++ DA +A+D
Sbjct 192 NVYIKNFGEDMDDERLKETFSKYGKTLSVKVMTDP-SGKSKGFGFVSFERHEDANKAVDD 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q R L+R N+++KNLD +ID++ L
Sbjct 251 MNGKDVNGKIMFVGRAQKKVERQAELKRRFEQLKQERISRYQGVNLYIKNLDDTIDDEKL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
FS FG+I S KV ++E RS+G+GFV + + A A+
Sbjct 311 RKEFSPFGSITSAKVMLEE-GRSKGFGFVCFSSPEEATKAV 350
Score = 37.4 bits (85), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 51/99 (51%), Gaps = 3/99 (3%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LYI +L + + L + F+ G +TS +V + RS G+ +V + S +A +
Sbjct 291 YQGVNLYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLE--EGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNL 108
A+ +N ++ +P + +QR R++ N +++ +
Sbjct 349 AVTEMNGRIVGSKPLYVALAQRKEE-RKAHLTNQYMQRI 386
> dre:321035 pabpc4, cb12, sb:cb12; poly(A) binding protein, cytoplasmic
4 (inducible form); K13126 polyadenylate-binding
protein
Length=637
Score = 235 bits (600), Expect = 4e-62, Method: Composition-based stats.
Identities = 111/159 (69%), Positives = 131/159 (82%), Gaps = 4/159 (2%)
Query 2 AGAGPHPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNY 61
A AG +P SLY+GDLHP++TEAML+E F+ GPV SIRVCRD ++RRSLGYAYVN+
Sbjct 4 ATAGSYPM---ASLYVGDLHPDITEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNF 60
Query 62 QSLGDAERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTF 121
Q DAERALD +NF +++G+P RIMWSQRDPSLR+SG GNVF+KNLDKSIDNKALYDTF
Sbjct 61 QQPADAERALDTMNFDVVKGKPIRIMWSQRDPSLRKSGVGNVFIKNLDKSIDNKALYDTF 120
Query 122 SLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
S FGNILSCKV DE N S+GY FVH+ET+ +A AIEK
Sbjct 121 SAFGNILSCKVVCDE-NGSKGYAFVHFETQDAADRAIEK 158
Score = 75.9 bits (185), Expect = 5e-14, Method: Composition-based stats.
Identities = 46/153 (30%), Positives = 80/153 (52%), Gaps = 8/153 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GYA+V++++ A+RA++
Sbjct 101 NVFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDE--NGSKGYAFVHFETQDAADRAIEK 158
Query 74 LNFALIRGQPCRI-MWSQRDPSLRRSGAG-----NVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + + R GA NV++KN +D++ L + F +G
Sbjct 159 MNGMLLNDRKVFVGRFKSRKEREAEMGAKAKEFTNVYIKNFGDDMDDQRLKELFDKYGKT 218
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LS KV D +SRG+GFV YE A A+E+
Sbjct 219 LSVKVMTDPTGKSRGFGFVSYEKHEDANKAVEE 251
Score = 72.8 bits (177), Expect = 5e-13, Method: Composition-based stats.
Identities = 47/161 (29%), Positives = 85/161 (52%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L E+F+ G S++V D + S G+ +V+Y+ DA +A++
Sbjct 193 NVYIKNFGDDMDDQRLKELFDKYGKTLSVKVMTDPTGK-SRGFGFVSYEKHEDANKAVEE 251
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q R L+R N+++KNLD +ID++ L
Sbjct 252 MNGTELNGKTVFVGRAQKKMERQAELKRKFEQLKQERISRYQGVNLYIKNLDDTIDDEKL 311
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
FS FG+I S KV ++E RS+G+GFV + + A A+
Sbjct 312 RKEFSPFGSITSAKVMLEE-GRSKGFGFVCFSSPEEATKAV 351
Score = 37.0 bits (84), Expect = 0.026, Method: Composition-based stats.
Identities = 22/82 (26%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LYI +L + + L + F+ G +TS +V + RS G+ +V + S +A +
Sbjct 292 YQGVNLYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLE--EGRSKGFGFVCFSSPEEATK 349
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 350 AVTEMNGRIVGSKPLYVALAQR 371
> bbo:BBOV_IV008280 23.m06515; polyadenylate binding protein;
K13126 polyadenylate-binding protein
Length=585
Score = 235 bits (599), Expect = 6e-62, Method: Compositional matrix adjust.
Identities = 107/152 (70%), Positives = 127/152 (83%), Gaps = 0/152 (0%)
Query 9 TYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAE 68
++ S SLY+G+L PEVTEAML+EVFN +GPV SIRVCRDS++R+SLGYAYVNY + DAE
Sbjct 32 SFNSASLYVGNLLPEVTEAMLYEVFNGIGPVASIRVCRDSLTRKSLGYAYVNYYNFQDAE 91
Query 69 RALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNIL 128
AL+ LN+ I+GQP RIMWS+RDPSLR+SG GN+FVKNLDK+ID KALYDTFS FG IL
Sbjct 92 AALECLNYIEIKGQPARIMWSERDPSLRKSGTGNIFVKNLDKAIDTKALYDTFSHFGTIL 151
Query 129 SCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
SCKV +D S+GYGFVHY TE SA+ AIEK
Sbjct 152 SCKVAIDSLGNSKGYGFVHYTTEESAKEAIEK 183
Score = 55.1 bits (131), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 42/154 (27%), Positives = 76/154 (49%), Gaps = 16/154 (10%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++++ +L + L++ F+ G + S +V DS+ S GY +V+Y + A+ A++
Sbjct 125 NIFVKNLDKAIDTKALYDTFSHFGTILSCKVAIDSLGN-SKGYGFVHYTTEESAKEAIEK 183
Query 74 LNFALIRGQPCRIMWSQRDPSLRR----SGAGNVF----VKNLDKSIDNKALYDTFSLFG 125
+N LI + P LRR S G+VF V+N + L+ TFS +G
Sbjct 184 VNGMLIGNSQVSVA-----PFLRRNERTSTVGDVFTNLYVRNFPDTWTEDDLHQTFSKYG 238
Query 126 NILSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
I S + D+ + R + FV++ A++A+E
Sbjct 239 EITSLLLKSDD--KGRRFAFVNFVDTNMAKAAME 270
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 44/167 (26%), Positives = 73/167 (43%), Gaps = 29/167 (17%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+ + TE L + F+ G +TS+ + D RR +A+VN+ A+ A++
Sbjct 215 NLYVRNFPDTWTEDDLHQTFSKYGEITSLLLKSDDKGRR---FAFVNFVDTNMAKAAMEG 271
Query 74 LN----------------------FALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKS 111
N +A+++ Q QR+ + N+++KNLD
Sbjct 272 ENGVKFESVEEPMMVCQHMDKARRYAMLKAQYDSNAQDQRNKFM----GVNLYIKNLDDD 327
Query 112 IDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
D+ L D F +G + S KV D SRG+GFV + A A+
Sbjct 328 FDDDGLRDLFKQYGTVTSSKVMRDHNGVSRGFGFVCFSRPDEATKAV 374
Score = 31.6 bits (70), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 46/84 (54%), Gaps = 5/84 (5%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRD--SVSRRSLGYAYVNYQSLGDA 67
++ +LYI +L + + L ++F G VTS +V RD VSR G+ +V + +A
Sbjct 314 FMGVNLYIKNLDDDFDDDGLRDLFKQYGTVTSSKVMRDHNGVSR---GFGFVCFSRPDEA 370
Query 68 ERALDALNFALIRGQPCRIMWSQR 91
+A+ ++ L++ +P + +++
Sbjct 371 TKAVAGMHLKLVKNKPLYVGLAEK 394
> xla:379896 pabpc1-a, MGC53109, pab1, pabp, pabp1, pabpc1, pabpc2;
poly(A) binding protein, cytoplasmic 1; K13126 polyadenylate-binding
protein
Length=633
Score = 235 bits (599), Expect = 6e-62, Method: Composition-based stats.
Identities = 108/153 (70%), Positives = 129/153 (84%), Gaps = 1/153 (0%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
P+Y SLY+GDLH +VTEAML+E F+ GP+ SIRVCRD ++RRSLGYAYVN+Q DA
Sbjct 6 PSYPMASLYVGDLHQDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADA 65
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
ERALD +NF +I+G+P RIMWSQRDPSLR+SG GN+F+KNLDKSIDNKALYDTFS FGNI
Sbjct 66 ERALDTMNFDVIKGRPVRIMWSQRDPSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNI 125
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LSCKV DE N S+GYGFVH+ET+ +A AI+K
Sbjct 126 LSCKVVCDE-NGSKGYGFVHFETQEAAERAIDK 157
Score = 71.2 bits (173), Expect = 1e-12, Method: Composition-based stats.
Identities = 43/153 (28%), Positives = 82/153 (53%), Gaps = 8/153 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GY +V++++ AERA+D
Sbjct 100 NIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDE--NGSKGYGFVHFETQEAAERAIDK 157
Query 74 LNFALIRGQPCRI----MWSQRDPSL--RRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + +R+ L R NV++KN ++++ L + F +G
Sbjct 158 MNGMLLNDRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGDDMNDERLKEMFGKYGPA 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LS KV D+ +S+G+GFV +E A+ A+++
Sbjct 218 LSVKVMTDDNGKSKGFGFVSFERHEDAQKAVDE 250
Score = 66.2 bits (160), Expect = 4e-11, Method: Composition-based stats.
Identities = 47/161 (29%), Positives = 82/161 (50%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L E+F GP S++V D + +S G+ +V+++ DA++A+D
Sbjct 192 NVYIKNFGDDMNDERLKEMFGKYGPALSVKVMTDD-NGKSKGFGFVSFERHEDAQKAVDE 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+ + G+ + +Q R L+R N++VKNLD ID++ L
Sbjct 251 MYGKDMNGKSMFVGRAQKKVERQTELKRKFEQMNQDRITRYQGVNLYVKNLDDGIDDERL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
F FG I S KV + E RS+G+GFV + + A A+
Sbjct 311 RKEFLPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAV 350
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 92 DPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFN-RSRGYGFVHYET 150
+PS +++V +L + + LY+ FS G ILS +V D RS GY +V+++
Sbjct 2 NPSAPSYPMASLYVGDLHQDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQ 61
Query 151 EASARSAIE 159
A A A++
Sbjct 62 PADAERALD 70
Score = 34.3 bits (77), Expect = 0.20, Method: Composition-based stats.
Identities = 21/82 (25%), Positives = 42/82 (51%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F G +TS +V + RS G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDGIDDERLRKEFLPFGTITSAKVMME--GGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 349 AVTEMNGRIVATKPLYVALAQR 370
> hsa:8761 PABPC4, APP-1, APP1, FLJ43938, PABP4, iPABP; poly(A)
binding protein, cytoplasmic 4 (inducible form); K13126 polyadenylate-binding
protein
Length=660
Score = 234 bits (597), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 109/152 (71%), Positives = 127/152 (83%), Gaps = 1/152 (0%)
Query 9 TYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAE 68
+Y SLY+GDLH +VTEAML+E F+ GPV SIRVCRD ++RRSLGYAYVN+Q DAE
Sbjct 7 SYPMASLYVGDLHSDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAE 66
Query 69 RALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNIL 128
RALD +NF +I+G+P RIMWSQRDPSLR+SG GNVF+KNLDKSIDNKALYDTFS FGNIL
Sbjct 67 RALDTMNFDVIKGKPIRIMWSQRDPSLRKSGVGNVFIKNLDKSIDNKALYDTFSAFGNIL 126
Query 129 SCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
SCKV DE N S+GY FVH+ET+ +A AIEK
Sbjct 127 SCKVVCDE-NGSKGYAFVHFETQEAADKAIEK 157
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 50/161 (31%), Positives = 87/161 (54%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + EV + L E+F+ G S++V RD + +S G+ +V+Y+ DA +A++
Sbjct 192 NVYIKNFGEEVDDESLKELFSQFGKTLSVKVMRDP-NGKSKGFGFVSYEKHEDANKAVEE 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N I G+ + +Q R L+R N+++KNLD +ID++ L
Sbjct 251 MNGKEISGKIIFVGRAQKKVERQAELKRKFEQLKQERISRYQGVNLYIKNLDDTIDDEKL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
FS FG+I S KV +++ RS+G+GFV + + A A+
Sbjct 311 RKEFSPFGSITSAKVMLED-GRSKGFGFVCFSSPEEATKAV 350
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 46/153 (30%), Positives = 83/153 (54%), Gaps = 8/153 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GYA+V++++ A++A++
Sbjct 100 NVFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENG--SKGYAFVHFETQEAADKAIEK 157
Query 74 LNFALIRGQPCRI-MWSQRDPSLRRSGAG-----NVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + + R GA NV++KN + +D+++L + FS FG
Sbjct 158 MNGMLLNDRKVFVGRFKSRKEREAELGAKAKEFTNVYIKNFGEEVDDESLKELFSQFGKT 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LS KV D +S+G+GFV YE A A+E+
Sbjct 218 LSVKVMRDPNGKSKGFGFVSYEKHEDANKAVEE 250
Score = 37.4 bits (85), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LYI +L + + L + F+ G +TS +V + RS G+ +V + S +A +
Sbjct 291 YQGVNLYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLE--DGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 349 AVTEMNGRIVGSKPLYVALAQR 370
> mmu:230721 Pabpc4, MGC11665, MGC6685; poly(A) binding protein,
cytoplasmic 4; K13126 polyadenylate-binding protein
Length=660
Score = 233 bits (595), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 108/152 (71%), Positives = 127/152 (83%), Gaps = 1/152 (0%)
Query 9 TYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAE 68
+Y SLY+GDLH +VTEAML+E F+ GPV SIRVCRD ++RRSLGYAYVN+Q DAE
Sbjct 7 SYPMASLYVGDLHSDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAE 66
Query 69 RALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNIL 128
RALD +NF +++G+P RIMWSQRDPSLR+SG GNVF+KNLDKSIDNKALYDTFS FGNIL
Sbjct 67 RALDTMNFDVMKGKPIRIMWSQRDPSLRKSGVGNVFIKNLDKSIDNKALYDTFSAFGNIL 126
Query 129 SCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
SCKV DE N S+GY FVH+ET+ +A AIEK
Sbjct 127 SCKVVCDE-NGSKGYAFVHFETQEAADKAIEK 157
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 51/161 (31%), Positives = 88/161 (54%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + EV + L E+F+ G S++V RDS S +S G+ +V+Y+ DA +A++
Sbjct 192 NVYIKNFGEEVDDGNLKELFSQFGKTLSVKVMRDS-SGKSKGFGFVSYEKHEDANKAVEE 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q R L+R N+++KNLD +ID++ L
Sbjct 251 MNGKEMSGKAIFVGRAQKKVERQAELKRKFEQLKQERISRYQGVNLYIKNLDDTIDDEKL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
FS FG+I S KV +++ RS+G+GFV + + A A+
Sbjct 311 RREFSPFGSITSAKVMLED-GRSKGFGFVCFSSPEEATKAV 350
Score = 75.5 bits (184), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 46/153 (30%), Positives = 81/153 (52%), Gaps = 8/153 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GYA+V++++ A++A++
Sbjct 100 NVFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENG--SKGYAFVHFETQEAADKAIEK 157
Query 74 LNFALIRGQPCRI-MWSQRDPSLRRSGAG-----NVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + + R GA NV++KN + +D+ L + FS FG
Sbjct 158 MNGMLLNDRKVFVGRFKSRKEREAELGAKAKEFTNVYIKNFGEEVDDGNLKELFSQFGKT 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LS KV D +S+G+GFV YE A A+E+
Sbjct 218 LSVKVMRDSSGKSKGFGFVSYEKHEDANKAVEE 250
Score = 36.6 bits (83), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 42/82 (51%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LYI +L + + L F+ G +TS +V + RS G+ +V + S +A +
Sbjct 291 YQGVNLYIKNLDDTIDDEKLRREFSPFGSITSAKVMLE--DGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 349 AVTEMNGRIVGSKPLYVALAQR 370
> dre:393856 pabpc1b, MGC77608, pabpc1, zgc:77608; poly A binding
protein, cytoplasmic 1 b; K13126 polyadenylate-binding protein
Length=634
Score = 233 bits (594), Expect = 2e-61, Method: Composition-based stats.
Identities = 108/153 (70%), Positives = 128/153 (83%), Gaps = 1/153 (0%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
P+Y SLY+GDLH +VTEAML+E F+ G + SIRVCRD ++RRSLGYAYVN+Q DA
Sbjct 6 PSYPMASLYVGDLHQDVTEAMLYEKFSPAGAILSIRVCRDMITRRSLGYAYVNFQQPADA 65
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
ERALD +NF +I+G+P RIMWSQRDPSLR+SG GN+F+KNLDKSIDNKALYDTFS FGNI
Sbjct 66 ERALDTMNFDVIKGRPVRIMWSQRDPSLRKSGVGNIFIKNLDKSIDNKALYDTFSAFGNI 125
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LSCKV DE N S+GYGFVH+ET+ +A AIEK
Sbjct 126 LSCKVVCDE-NGSKGYGFVHFETQEAAERAIEK 157
Score = 79.0 bits (193), Expect = 6e-15, Method: Composition-based stats.
Identities = 46/153 (30%), Positives = 84/153 (54%), Gaps = 8/153 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GY +V++++ AERA++
Sbjct 100 NIFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDE--NGSKGYGFVHFETQEAAERAIEK 157
Query 74 LNFALIRGQPCRI----MWSQRDPSL--RRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + +R+ L R NV++KN + +D+ L D FS +GN
Sbjct 158 MNGMLLNDRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGEDMDDDKLKDIFSKYGNA 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
+S +V DE +SRG+GFV +E A+ A+++
Sbjct 218 MSIRVMTDENGKSRGFGFVSFERHEDAQRAVDE 250
Score = 72.0 bits (175), Expect = 8e-13, Method: Composition-based stats.
Identities = 50/161 (31%), Positives = 84/161 (52%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L ++F+ G SIRV D + +S G+ +V+++ DA+RA+D
Sbjct 192 NVYIKNFGEDMDDDKLKDIFSKYGNAMSIRVMTDE-NGKSRGFGFVSFERHEDAQRAVDE 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q R L+R N++VKNLD ID++ L
Sbjct 251 MNGKEMNGKLIYVGRAQKKVERQTELKRKFEQMKQDRMTRYQGVNLYVKNLDDGIDDERL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
FS FG I S KV +D RS+G+GFV + + A A+
Sbjct 311 RKEFSPFGTITSAKVMMDG-GRSKGFGFVCFSSPEEATKAV 350
Score = 38.1 bits (87), Expect = 0.011, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 92 DPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFN-RSRGYGFVHYET 150
+PS +++V +L + + LY+ FS G ILS +V D RS GY +V+++
Sbjct 2 NPSAPSYPMASLYVGDLHQDVTEAMLYEKFSPAGAILSIRVCRDMITRRSLGYAYVNFQQ 61
Query 151 EASARSAIE 159
A A A++
Sbjct 62 PADAERALD 70
Score = 37.4 bits (85), Expect = 0.019, Method: Composition-based stats.
Identities = 22/82 (26%), Positives = 43/82 (52%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F+ G +TS +V D RS G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDGIDDERLRKEFSPFGTITSAKVMMD--GGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 349 AVTEMNGRIVATKPLYVALAQR 370
> tpv:TP01_0945 polyadenylate binding protein; K13126 polyadenylate-binding
protein
Length=661
Score = 232 bits (591), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 110/151 (72%), Positives = 124/151 (82%), Gaps = 0/151 (0%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
+ S SLY+GDL P+VTEA+L+EVFN VGPV SIRVCRDSV+R+SLGYAYVNY S DA+
Sbjct 24 FSSASLYVGDLKPDVTEAVLYEVFNTVGPVASIRVCRDSVTRKSLGYAYVNYYSTQDAQE 83
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
AL+ LN+ I+G P RIMWS RDPSLRRSGAGN+FVKNLDKSID K+LYDTFS FG ILS
Sbjct 84 ALENLNYIEIKGHPTRIMWSNRDPSLRRSGAGNIFVKNLDKSIDTKSLYDTFSHFGPILS 143
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
CKV VD S+ YGFVHYE E SAR AIEK
Sbjct 144 CKVAVDASGASKRYGFVHYENEESAREAIEK 174
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 49/167 (29%), Positives = 76/167 (45%), Gaps = 29/167 (17%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+ + + E L + G +TS+ + DS RR +A+VNY+ A+ ++
Sbjct 204 NLYVRNFPADWDEEALRQFLEKYGEITSMMLKEDSKGRR---FAFVNYKEPEVAKEVVNT 260
Query 74 LN----------------------FALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKS 111
LN L+R Q +Q D + N+++KNLD S
Sbjct 261 LNDLKLEESSEPLLVCPHQDKAKRQNLLRAQFNNSTMAQED----KRVTSNLYIKNLDDS 316
Query 112 IDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
D+++L + F FG I S KV +D N SRG+GFV + A AI
Sbjct 317 FDDESLGELFKPFGTITSSKVMLDANNHSRGFGFVCFTNPQEATKAI 363
> mmu:100503296 polyadenylate-binding protein 4-like
Length=234
Score = 232 bits (591), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 108/152 (71%), Positives = 127/152 (83%), Gaps = 1/152 (0%)
Query 9 TYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAE 68
+Y SLY+GDLH +VTEAML+E F+ GPV SIRVCRD ++RRSLGYAYVN+Q DAE
Sbjct 7 SYPMASLYVGDLHSDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAE 66
Query 69 RALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNIL 128
RALD +NF +++G+P RIMWSQRDPSLR+SG GNVF+KNLDKSIDNKALYDTFS FGNIL
Sbjct 67 RALDTMNFDVMKGKPIRIMWSQRDPSLRKSGVGNVFIKNLDKSIDNKALYDTFSAFGNIL 126
Query 129 SCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
SCKV DE N S+GY FVH+ET+ +A AIEK
Sbjct 127 SCKVVCDE-NGSKGYAFVHFETQEAADKAIEK 157
Score = 62.0 bits (149), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 39/137 (28%), Positives = 72/137 (52%), Gaps = 8/137 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D S GYA+V++++ A++A++
Sbjct 100 NVFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDE--NGSKGYAFVHFETQEAADKAIEK 157
Query 74 LNFALIRGQPCRI-MWSQRDPSLRRSGAG-----NVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + + R GA NV++KN + +D+ L + FS FG
Sbjct 158 MNGMLLNDRKVFVGRFKSRKEREAELGAKAKEFTNVYIKNFGEEVDDGNLKELFSQFGKT 217
Query 128 LSCKVGVDEFNRSRGYG 144
LS KV D +S+G+G
Sbjct 218 LSVKVMRDSSGKSKGFG 234
> mmu:67543 Pabpc6, 4932702K14Rik, AI428050, MGC132900, Pabpc3;
poly(A) binding protein, cytoplasmic 6; K13126 polyadenylate-binding
protein
Length=643
Score = 232 bits (591), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 107/153 (69%), Positives = 127/153 (83%), Gaps = 1/153 (0%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
P+Y SLY+GDLHP+VTEAML+E F+ GP+ SIRV RD ++RRSLGYA VN+Q L DA
Sbjct 6 PSYSLASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVYRDRITRRSLGYASVNFQQLEDA 65
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
ERALD +NF +I+G+P RIMWSQRDPSLR+SG GN+FVKNLD+SID+K LYDTFS FGNI
Sbjct 66 ERALDTMNFDVIKGKPVRIMWSQRDPSLRKSGVGNIFVKNLDRSIDSKTLYDTFSAFGNI 125
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LSCKV DE N S+GYGFVH+ET+ A AIEK
Sbjct 126 LSCKVVCDE-NGSKGYGFVHFETQEEAERAIEK 157
Score = 82.4 bits (202), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 49/156 (31%), Positives = 86/156 (55%), Gaps = 14/156 (8%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++++ +L + L++ F+A G + S +V D S GY +V++++ +AERA++
Sbjct 100 NIFVKNLDRSIDSKTLYDTFSAFGNILSCKVVCDENG--SKGYGFVHFETQEEAERAIEK 157
Query 74 LNFALIRGQPCRI--MWSQRDPSLRRSGAG-------NVFVKNLDKSIDNKALYDTFSLF 124
+N + + S+RD R++ G NV++KNL + +D++ L D F F
Sbjct 158 MNGMFLNDHKVFVGRFKSRRD---RQAELGARAKEFTNVYIKNLGEDMDDERLQDLFGRF 214
Query 125 GNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
G LS KV DE +S+G+GFV +E AR A+E+
Sbjct 215 GPALSVKVMTDESGKSKGFGFVSFERHEDARKAVEE 250
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 50/172 (29%), Positives = 85/172 (49%), Gaps = 30/172 (17%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI +L ++ + L ++F GP S++V D S +S G+ +V+++ DA +A++
Sbjct 192 NVYIKNLGEDMDDERLQDLFGRFGPALSVKVMTDE-SGKSKGFGFVSFERHEDARKAVEE 250
Query 74 LN---------------------------FALIRGQPCRIMWSQRDPSLRRSGAGNVFVK 106
+N F ++ +I +D S+R G N++VK
Sbjct 251 MNGKDLNGKQIYVGRAQKKVERQTELKHKFGQMKQDKHKIERVPQDRSVRCKGV-NLYVK 309
Query 107 NLDKSIDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
NLD ID++ L FS FG I S KV + E RS+G+GFV + + A A+
Sbjct 310 NLDDGIDDERLRKEFSPFGTITSAKVTM-EGGRSKGFGFVCFSSPEEATKAV 360
> xla:443777 pabpc1l-b, MGC81363, epab, epabp-b, pabpc1l1; poly(A)
binding protein, cytoplasmic 1-like; K13126 polyadenylate-binding
protein
Length=629
Score = 229 bits (584), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 105/150 (70%), Positives = 124/150 (82%), Gaps = 1/150 (0%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y SLYIGDLHP+VTEAML+E F+ GP+ SIRVCRD +RRSLGYAY+N+Q DAER
Sbjct 8 YPLASLYIGDLHPDVTEAMLYEKFSPAGPIMSIRVCRDIATRRSLGYAYINFQQPADAER 67
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
ALD +NF +I+G+P RIMWSQRDP LR+SG GNVF+KNLD SIDNKALYDTFS FG+ILS
Sbjct 68 ALDTMNFEVIKGRPIRIMWSQRDPGLRKSGVGNVFIKNLDDSIDNKALYDTFSAFGDILS 127
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAIE 159
CKV DE+ SRGYGFVH+ET+ +A AI+
Sbjct 128 CKVVCDEYG-SRGYGFVHFETQEAANRAIQ 156
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/151 (31%), Positives = 81/151 (53%), Gaps = 8/151 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D R GY +V++++ A RA+
Sbjct 100 NVFIKNLDDSIDNKALYDTFSAFGDILSCKVVCDEYGSR--GYGFVHFETQEAANRAIQT 157
Query 74 LNFALIRGQPCRI--MWSQRDPSLRRSGA----GNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + S+R+ L NV++KN + +D+K L + FS FGN
Sbjct 158 MNGMLLNDRKVFVGHFKSRRERELEYGAKVMEFTNVYIKNFGEDMDDKRLKEIFSAFGNT 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
LS KV +D RSRG+GFV+Y A+ A+
Sbjct 218 LSVKVMMDNSGRSRGFGFVNYGNHEEAQKAV 248
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 47/161 (29%), Positives = 82/161 (50%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L E+F+A G S++V D+ S RS G+ +VNY + +A++A+
Sbjct 192 NVYIKNFGEDMDDKRLKEIFSAFGNTLSVKVMMDN-SGRSRGFGFVNYGNHEEAQKAVTE 250
Query 74 LNFALIRGQPCRIMWSQR----------------DPSLRRSGAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q+ + R N++VKNLD ID+ L
Sbjct 251 MNGKEVNGRMVYVGRAQKRIERQGELKRKFEQIKQERINRYQGVNLYVKNLDDGIDDDRL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
FS +G I S KV + E S+G+GFV + + A A+
Sbjct 311 RKEFSPYGTITSTKV-MTEGGHSKGFGFVCFSSPEEATKAV 350
Score = 33.5 bits (75), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 23/99 (23%), Positives = 50/99 (50%), Gaps = 3/99 (3%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F+ G +TS +V + S G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDGIDDDRLRKEFSPYGTITSTKVMTE--GGHSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNL 108
A+ +N ++ +P + +QR R++ N +++ L
Sbjct 349 AVTEMNGRIVSTKPLYVALAQRKEE-RKAILTNQYMQRL 386
> xla:398221 pabpc1l-a, MGC82273, ePAB, epabp-a, pabpc1l1; poly(A)
binding protein, cytoplasmic 1-like; K13126 polyadenylate-binding
protein
Length=629
Score = 228 bits (581), Expect = 7e-60, Method: Compositional matrix adjust.
Identities = 108/158 (68%), Positives = 125/158 (79%), Gaps = 5/158 (3%)
Query 2 AGAGPHPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNY 61
GAG Y SLYIGDLHP+VTEAML+E F+ GP+ SIRVCRD +RRSL YAY+N+
Sbjct 4 TGAG----YPLASLYIGDLHPDVTEAMLYEKFSPAGPIMSIRVCRDIATRRSLSYAYINF 59
Query 62 QSLGDAERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTF 121
Q DAERALD +NF +I+G+P RIMWSQRDP LR+SG GNVF+KNLD+SIDNKALYDTF
Sbjct 60 QQPADAERALDTMNFEVIKGRPIRIMWSQRDPGLRKSGVGNVFIKNLDESIDNKALYDTF 119
Query 122 SLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
S FGNILSCKV DE SRGYGFVH+ET +A AI+
Sbjct 120 SAFGNILSCKVVCDEHG-SRGYGFVHFETHEAANRAIQ 156
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/151 (31%), Positives = 82/151 (54%), Gaps = 8/151 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+A G + S +V D R GY +V++++ A RA+
Sbjct 100 NVFIKNLDESIDNKALYDTFSAFGNILSCKVVCDEHGSR--GYGFVHFETHEAANRAIQT 157
Query 74 LNFALIRGQPCRI--MWSQRDPSLRRSGA----GNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + S+R+ L NV++KN + +D+K L + FS FGN
Sbjct 158 MNGMLLNDRKVFVGHFKSRRERELEYGAKVMEFTNVYIKNFGEDMDDKRLREIFSAFGNT 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
LS KV +D+ RSRG+GFV+Y A+ A+
Sbjct 218 LSVKVMMDDSGRSRGFGFVNYGNHEEAQKAV 248
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 48/161 (29%), Positives = 81/161 (50%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L E+F+A G S++V D S RS G+ +VNY + +A++A+
Sbjct 192 NVYIKNFGEDMDDKRLREIFSAFGNTLSVKVMMDD-SGRSRGFGFVNYGNHEEAQKAVSE 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q R L+R N++VKNLD ID+ L
Sbjct 251 MNGKEVNGRMIYVGRAQKRIERQSELKRKFEQIKQERINRYQGVNLYVKNLDDGIDDDRL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
F +G I S KV + E S+G+GFV + + A A+
Sbjct 311 RKEFLPYGTITSAKV-MTEGGHSKGFGFVCFSSPEEATKAV 350
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/99 (23%), Positives = 49/99 (49%), Gaps = 3/99 (3%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F G +TS +V + S G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDGIDDDRLRKEFLPYGTITSAKVMTE--GGHSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNL 108
A+ +N ++ +P + +QR R++ N +++ L
Sbjct 349 AVTEMNGRIVSTKPLYVALAQRKEE-RKAILTNQYMQRL 386
> mmu:18459 Pabpc2, PABP, PABP+, Pabp2; poly(A) binding protein,
cytoplasmic 2; K13126 polyadenylate-binding protein
Length=628
Score = 228 bits (580), Expect = 9e-60, Method: Compositional matrix adjust.
Identities = 105/147 (71%), Positives = 126/147 (85%), Gaps = 1/147 (0%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SLY+GDLHP+VTEAML+E F++ GP+ SIRV RD ++RRSLGYA VN++ DAERALD
Sbjct 12 SLYVGDLHPDVTEAMLYEKFSSAGPILSIRVYRDVITRRSLGYASVNFEQPADAERALDT 71
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVG 133
+NF +I+G+P RIMWSQRDPSLRRSG GNVF+KNL+K+IDNKALYDTFS FGNILSCKV
Sbjct 72 MNFDVIKGKPVRIMWSQRDPSLRRSGVGNVFIKNLNKTIDNKALYDTFSAFGNILSCKVV 131
Query 134 VDEFNRSRGYGFVHYETEASARSAIEK 160
DE N S+G+GFVH+ETE +A AIEK
Sbjct 132 SDE-NGSKGHGFVHFETEEAAERAIEK 157
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 46/156 (29%), Positives = 84/156 (53%), Gaps = 14/156 (8%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L+ + L++ F+A G + S +V D S G+ +V++++ AERA++
Sbjct 100 NVFIKNLNKTIDNKALYDTFSAFGNILSCKVVSDE--NGSKGHGFVHFETEEAAERAIEK 157
Query 74 LNFALIRGQPCRI----MWSQRDPSLRRSGAG-----NVFVKNLDKSIDNKALYDTFSLF 124
+N L+ + + +R+ L G G NV++KN +D++ L F F
Sbjct 158 MNGMLLNDRKVFVGRFKSQKEREAEL---GTGTKEFTNVYIKNFGDRMDDETLNGLFGRF 214
Query 125 GNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
G ILS KV DE +S+G+GFV +E A+ A+++
Sbjct 215 GQILSVKVMTDEGGKSKGFGFVSFERHEDAQKAVDE 250
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 48/164 (29%), Positives = 84/164 (51%), Gaps = 20/164 (12%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + + + L +F G + S++V D +S G+ +V+++ DA++A+D
Sbjct 192 NVYIKNFGDRMDDETLNGLFGRFGQILSVKVMTDE-GGKSKGFGFVSFERHEDAQKAVDE 250
Query 74 LNFALIRGQPCRIMWSQR-----------------DPSLRRSGAGNVFVKNLDKSIDNKA 116
+N + G+ + +Q+ D S+R G N++VKNLD ID++
Sbjct 251 MNGKELNGKHIYVGRAQKKDDRHTELKHKFEQVTQDKSIRYQGI-NLYVKNLDDGIDDER 309
Query 117 LYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
L FS FG I S KV + E RS+G+GFV + + A A+ +
Sbjct 310 LQKEFSPFGTITSTKV-MTEGGRSKGFGFVCFSSPEEATKAVSE 352
Score = 37.4 bits (85), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 23/99 (23%), Positives = 51/99 (51%), Gaps = 3/99 (3%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F+ G +TS +V + RS G+ +V + S +A +
Sbjct 291 YQGINLYVKNLDDGIDDERLQKEFSPFGTITSTKVMTE--GGRSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNL 108
A+ +N ++ +P + +QR R++ N +++ +
Sbjct 349 AVSEMNGRIVATKPLYVALAQRKEE-RQAHLTNQYIQRM 386
> hsa:80336 PABPC1L, C20orf119, FLJ11840, FLJ30809, FLJ42053,
PABPC1L1, dJ1069P2.3, ePAB; poly(A) binding protein, cytoplasmic
1-like; K13126 polyadenylate-binding protein
Length=614
Score = 227 bits (579), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 102/149 (68%), Positives = 122/149 (81%), Gaps = 1/149 (0%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y SLY+GDLHP+VTEAML+E F+ GP+ SIRVCRD +RRSLGYAY+N+Q DAER
Sbjct 8 YPLASLYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDVATRRSLGYAYINFQQPADAER 67
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
ALD +NF +++GQP RIMWSQRDP LR+SG GN+F+KNL+ SIDNKALYDTFS FGNILS
Sbjct 68 ALDTMNFEMLKGQPIRIMWSQRDPGLRKSGVGNIFIKNLEDSIDNKALYDTFSTFGNILS 127
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAI 158
CKV DE SRG+GFVH+ET +A+ AI
Sbjct 128 CKVACDEHG-SRGFGFVHFETHEAAQQAI 155
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/151 (28%), Positives = 79/151 (52%), Gaps = 8/151 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+ G + S +V D R G+ +V++++ A++A++
Sbjct 100 NIFIKNLEDSIDNKALYDTFSTFGNILSCKVACDEHGSR--GFGFVHFETHEAAQQAINT 157
Query 74 LNFALIRGQPCRI----MWSQRDPSL--RRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + +R+ L R N++VKNL +D + L D FS FG +
Sbjct 158 MNGMLLNDRKVFVGHFKSRREREAELGARALEFTNIYVKNLPVDVDEQGLQDLFSQFGKM 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
LS KV D SR +GFV++E A+ A+
Sbjct 218 LSVKVMRDNSGHSRCFGFVNFEKHEEAQKAV 248
Score = 69.3 bits (168), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 49/164 (29%), Positives = 85/164 (51%), Gaps = 24/164 (14%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+ +L +V E L ++F+ G + S++V RD+ S S + +VN++ +A++A+
Sbjct 192 NIYVKNLPVDVDEQGLQDLFSQFGKMLSVKVMRDN-SGHSRCFGFVNFEKHEEAQKAVVH 250
Query 74 LNFALIRGQPCRIMWSQR-------------------DPSLRRSGAGNVFVKNLDKSIDN 114
+N + G R++++ R LRR N++VKNLD SID+
Sbjct 251 MNGKEVSG---RLLYAGRAQKRVERQNELKRRFEQMKQDRLRRYQGVNLYVKNLDDSIDD 307
Query 115 KALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
L FS +G I S KV + E S+G+GFV + + A A+
Sbjct 308 DKLRKEFSPYGVITSAKV-MTEGGHSKGFGFVCFSSPEEATKAV 350
Score = 33.1 bits (74), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 23/99 (23%), Positives = 50/99 (50%), Gaps = 3/99 (3%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F+ G +TS +V + S G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDSIDDDKLRKEFSPYGVITSAKVMTE--GGHSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNL 108
A+ +N ++ +P + +QR R++ N +++ L
Sbjct 349 AVTEMNGRIVGTKPLYVALAQRKEE-RKAILTNQYMQRL 386
> dre:327625 pabpc1l, fi20g03, pab-2, wu:fi20g03, zgc:55855; poly(A)
binding protein, cytoplasmic 1-like; K13126 polyadenylate-binding
protein
Length=620
Score = 221 bits (563), Expect = 7e-58, Method: Compositional matrix adjust.
Identities = 100/152 (65%), Positives = 124/152 (81%), Gaps = 1/152 (0%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
P Y SLY+GDLH +VTEAML++ F+ G + SIRVCRD ++RRSLGYAY+N+Q DA
Sbjct 6 PAYPLASLYVGDLHADVTEAMLYQKFSPAGQIMSIRVCRDVITRRSLGYAYINFQQPADA 65
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
E ALD +N+ +I+G+P RIMWSQRDP LR+SG GN+F+KN+D+SIDNKALYDTFS FGNI
Sbjct 66 ECALDTMNYEVIKGRPIRIMWSQRDPGLRKSGVGNIFIKNMDESIDNKALYDTFSAFGNI 125
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
LSCKV DE N S+GYGFVH+ET+ +A AIE
Sbjct 126 LSCKVVCDE-NGSKGYGFVHFETQEAANRAIE 156
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 45/151 (29%), Positives = 79/151 (52%), Gaps = 8/151 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I ++ + L++ F+A G + S +V D S GY +V++++ A RA++
Sbjct 100 NIFIKNMDESIDNKALYDTFSAFGNILSCKVVCDENG--SKGYGFVHFETQEAANRAIET 157
Query 74 LNFALIRGQPCRI-MWSQRDPSLRRSGAG-----NVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + + R GA NV++KN + ID++ L + F+ FG
Sbjct 158 MNGMLLNDRKVFVGHFKSRKEREAEMGAKAVEFTNVYIKNFGEDIDSEKLKNIFTEFGKT 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
LS V DE RSRG+GFV++ AR A+
Sbjct 218 LSVCVMTDERGRSRGFGFVNFVNHGDARRAV 248
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 47/161 (29%), Positives = 77/161 (47%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ L +F G S+ V D R S G+ +VN+ + GDA RA+
Sbjct 192 NVYIKNFGEDIDSEKLKNIFTEFGKTLSVCVMTDERGR-SRGFGFVNFVNHGDARRAVTE 250
Query 74 LNFALIRGQPCRIMWSQR----------------DPSLRRSGAGNVFVKNLDKSIDNKAL 117
+N + G+ + +Q+ ++R N++VKNLD SID++ L
Sbjct 251 MNGKELNGRVLYVGRAQKRLERQGELKRKFEQIKQERIQRYQGVNLYVKNLDDSIDDEKL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
F+ +G I S KV D SRG+GFV + + A A+
Sbjct 311 RKEFAPYGTITSAKVMTDG-GHSRGFGFVCFSSPEEATKAV 350
Score = 37.4 bits (85), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 27/115 (23%), Positives = 56/115 (48%), Gaps = 5/115 (4%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L + F G +TS +V D R G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDSIDDEKLRKEFAPYGTITSAKVMTDGGHSR--GFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLD--KSIDNKALYDTFS 122
A+ +N ++ +P + +QR R++ N +++ L ++I A+ T+
Sbjct 349 AVTEMNGRIVSTKPLYVALAQRKEE-RKAILTNQYIQRLASIRAIPGPAIPTTYQ 402
> hsa:5042 PABPC3, PABP3, PABPL3, tPABP; poly(A) binding protein,
cytoplasmic 3; K13126 polyadenylate-binding protein
Length=631
Score = 221 bits (562), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 101/153 (66%), Positives = 123/153 (80%), Gaps = 1/153 (0%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
P+Y + SLY+GDLHP+VTEAML+E F+ GP+ SIR+CRD ++ S YAYVN+Q DA
Sbjct 6 PSYPTASLYVGDLHPDVTEAMLYEKFSPAGPILSIRICRDLITSGSSNYAYVNFQHTKDA 65
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
E ALD +NF +I+G+P RIMWSQRDPSLR+SG GN+FVKNLDKSI+NKALYDT S FGNI
Sbjct 66 EHALDTMNFDVIKGKPVRIMWSQRDPSLRKSGVGNIFVKNLDKSINNKALYDTVSAFGNI 125
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LSC V DE N S+GYGFVH+ET +A AI+K
Sbjct 126 LSCNVVCDE-NGSKGYGFVHFETHEAAERAIKK 157
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 50/162 (30%), Positives = 86/162 (53%), Gaps = 18/162 (11%)
Query 13 PSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALD 72
P++YI + ++ + L ++F GP S++V D S +S G+ +V+++ DA++A+D
Sbjct 191 PNVYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDE-SGKSKGFGFVSFERHEDAQKAVD 249
Query 73 ALNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKA 116
+N + G+ + +Q R L+R+ N++VKNLD ID++
Sbjct 250 EMNGKELNGKQIYVGRAQKKVERQTELKRTFEQMKQDRITRYQVVNLYVKNLDDGIDDER 309
Query 117 LYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
L FS FG I S KV + E RS+G+GFV + + A A+
Sbjct 310 LRKAFSPFGTITSAKVMM-EGGRSKGFGFVCFSSPEEATKAV 350
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 45/153 (29%), Positives = 81/153 (52%), Gaps = 8/153 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++++ +L + L++ +A G + S V D S GY +V++++ AERA+
Sbjct 100 NIFVKNLDKSINNKALYDTVSAFGNILSCNVVCDENG--SKGYGFVHFETHEAAERAIKK 157
Query 74 LNFALIRGQPCRIMW----SQRDPSL--RRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ G+ + +R+ L R NV++KN + +D++ L D F FG
Sbjct 158 MNGMLLNGRKVFVGQFKSRKEREAELGARAKEFPNVYIKNFGEDMDDERLKDLFGKFGPA 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LS KV DE +S+G+GFV +E A+ A+++
Sbjct 218 LSVKVMTDESGKSKGFGFVSFERHEDAQKAVDE 250
Score = 36.2 bits (82), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 20/78 (25%), Positives = 42/78 (53%), Gaps = 2/78 (2%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+ +L + + L + F+ G +TS +V + RS G+ +V + S +A +A+
Sbjct 295 NLYVKNLDDGIDDERLRKAFSPFGTITSAKVMME--GGRSKGFGFVCFSSPEEATKAVTE 352
Query 74 LNFALIRGQPCRIMWSQR 91
+N ++ +P + +QR
Sbjct 353 MNGRIVATKPLYVALAQR 370
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 33/69 (47%), Gaps = 1/69 (1%)
Query 92 DPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFNR-SRGYGFVHYET 150
+PS +++V +L + LY+ FS G ILS ++ D S Y +V+++
Sbjct 2 NPSTPSYPTASLYVGDLHPDVTEAMLYEKFSPAGPILSIRICRDLITSGSSNYAYVNFQH 61
Query 151 EASARSAIE 159
A A++
Sbjct 62 TKDAEHALD 70
> cpv:cgd6_3010 poly(a)-binding protein fabm ; K13126 polyadenylate-binding
protein
Length=746
Score = 221 bits (562), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 99/150 (66%), Positives = 122/150 (81%), Gaps = 0/150 (0%)
Query 11 VSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERA 70
VS SLY+GDL +VTE ML+E+FN+V V+S+R+CRD+++RRSLGYAYVNY S+ DAERA
Sbjct 10 VSASLYVGDLDADVTETMLYEIFNSVAVVSSVRICRDALTRRSLGYAYVNYNSVADAERA 69
Query 71 LDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSC 130
LD LNF IRG+PCRIMW RDP+ RR+ GNVFVKNLDKSIDNK L+DTFSLFGNI+SC
Sbjct 70 LDTLNFTCIRGRPCRIMWCLRDPASRRNNDGNVFVKNLDKSIDNKTLFDTFSLFGNIMSC 129
Query 131 KVGVDEFNRSRGYGFVHYETEASARSAIEK 160
K+ D +S GYGF+H+E SA+ AI +
Sbjct 130 KIATDVEGKSLGYGFIHFEHADSAKEAISR 159
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 102 NVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
N++VKNL SI+ + L F FG + S + DE SRG+GFV + + A AI
Sbjct 364 NLYVKNLADSINEEDLRSMFEPFGTVSSVSIKTDESGVSRGFGFVSFLSPDEATKAI 420
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Query 7 HPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGD 66
H Y +LY+ +L + E L +F G V+S+ + D S S G+ +V++ S +
Sbjct 357 HQRYQGVNLYVKNLADSINEEDLRSMFEPFGTVSSVSIKTDE-SGVSRGFGFVSFLSPDE 415
Query 67 AERALDALNFALIRGQPCRIMWSQR 91
A +A+ ++ L+RG+P + +R
Sbjct 416 ATKAITEMHLKLVRGKPLYVGLHER 440
Score = 34.7 bits (78), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 39/66 (59%), Gaps = 3/66 (4%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+ + TE +L+++F G ++S+ + DS R + +VN+++ A+ A+ A
Sbjct 191 NVYVKHIPKSWTEDLLYKIFGVYGKISSLVLQSDSKGRP---FGFVNFENPDSAKAAVAA 247
Query 74 LNFALI 79
L+ AL+
Sbjct 248 LHNALV 253
> mmu:668382 Pabpc1l2b-ps, EG668382, Pabpc1l2b, Rbm32b; poly(A)
binding protein, cytoplasmic 1-like 2B, pseudogene; K13126
polyadenylate-binding protein
Length=229
Score = 218 bits (555), Expect = 6e-57, Method: Compositional matrix adjust.
Identities = 100/153 (65%), Positives = 124/153 (81%), Gaps = 1/153 (0%)
Query 7 HPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGD 66
+P + + SLY+GDLHPEVTE+ML+E F+ GP+ SIR+CRD V+RRSLGYAYVNYQ D
Sbjct 25 NPDFPTASLYVGDLHPEVTESMLYEKFSPAGPILSIRICRDKVTRRSLGYAYVNYQQPVD 84
Query 67 AERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGN 126
A+RAL+ +NF +I G+P RIMWSQRDPSLR+SG GNVF+KNL K+IDNKALY+ FS FGN
Sbjct 85 AKRALETMNFDVINGRPVRIMWSQRDPSLRKSGVGNVFIKNLGKTIDNKALYNIFSAFGN 144
Query 127 ILSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
ILSCKV DE +GYGFVH++ + SA AI+
Sbjct 145 ILSCKVACDE-KGPKGYGFVHFQKQESAERAID 176
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L+ +F+A G + S +V D + GY +V++Q AERA+DA
Sbjct 120 NVFIKNLGKTIDNKALYNIFSAFGNILSCKVACDEKGPK--GYGFVHFQKQESAERAIDA 177
Query 74 LN 75
LN
Sbjct 178 LN 179
> hsa:340529 PABPC1L2A, MGC168104, PABPC1L2B, RBM32A; poly(A)
binding protein, cytoplasmic 1-like 2A; K13126 polyadenylate-binding
protein
Length=200
Score = 218 bits (555), Expect = 7e-57, Method: Compositional matrix adjust.
Identities = 100/146 (68%), Positives = 121/146 (82%), Gaps = 1/146 (0%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SLY+GDLHPEVTEAML+E F+ GP+ SIR+CRD ++RRSLGYAYVNYQ DA+RAL+
Sbjct 3 SLYVGDLHPEVTEAMLYEKFSPAGPILSIRICRDKITRRSLGYAYVNYQQPVDAKRALET 62
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVG 133
LNF +I+G+P RIMWSQRDPSLR+SG GNVF+KNL K+IDNKALY+ FS FGNILSCKV
Sbjct 63 LNFDVIKGRPVRIMWSQRDPSLRKSGVGNVFIKNLGKTIDNKALYNIFSAFGNILSCKVA 122
Query 134 VDEFNRSRGYGFVHYETEASARSAIE 159
DE +GYGFVH++ + SA AI+
Sbjct 123 CDE-KGPKGYGFVHFQKQESAERAID 147
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L+ +F+A G + S +V D + GY +V++Q AERA+D
Sbjct 91 NVFIKNLGKTIDNKALYNIFSAFGNILSCKVACDEKGPK--GYGFVHFQKQESAERAIDV 148
Query 74 LN 75
+N
Sbjct 149 MN 150
> hsa:645974 PABPC1L2B, MGC168104, PABPC1L2A, RBM32B; poly(A)
binding protein, cytoplasmic 1-like 2B; K13126 polyadenylate-binding
protein
Length=200
Score = 218 bits (555), Expect = 7e-57, Method: Compositional matrix adjust.
Identities = 100/146 (68%), Positives = 121/146 (82%), Gaps = 1/146 (0%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SLY+GDLHPEVTEAML+E F+ GP+ SIR+CRD ++RRSLGYAYVNYQ DA+RAL+
Sbjct 3 SLYVGDLHPEVTEAMLYEKFSPAGPILSIRICRDKITRRSLGYAYVNYQQPVDAKRALET 62
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVG 133
LNF +I+G+P RIMWSQRDPSLR+SG GNVF+KNL K+IDNKALY+ FS FGNILSCKV
Sbjct 63 LNFDVIKGRPVRIMWSQRDPSLRKSGVGNVFIKNLGKTIDNKALYNIFSAFGNILSCKVA 122
Query 134 VDEFNRSRGYGFVHYETEASARSAIE 159
DE +GYGFVH++ + SA AI+
Sbjct 123 CDE-KGPKGYGFVHFQKQESAERAID 147
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 35/62 (56%), Gaps = 2/62 (3%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L+ +F+A G + S +V D + GY +V++Q AERA+D
Sbjct 91 NVFIKNLGKTIDNKALYNIFSAFGNILSCKVACDEKGPK--GYGFVHFQKQESAERAIDV 148
Query 74 LN 75
+N
Sbjct 149 MN 150
> mmu:381404 Pabpc1l, 1810053B01Rik, Epab; poly(A) binding protein,
cytoplasmic 1-like; K13126 polyadenylate-binding protein
Length=607
Score = 214 bits (546), Expect = 8e-56, Method: Compositional matrix adjust.
Identities = 96/145 (66%), Positives = 120/145 (82%), Gaps = 1/145 (0%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SLY+GDLHP+VTE+ML+E+F+ +G + SIRVCRD +RRSLGYAY+N+Q DAERALD
Sbjct 12 SLYVGDLHPDVTESMLYEMFSPIGNILSIRVCRDVATRRSLGYAYINFQQPADAERALDT 71
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVG 133
+NF +I+GQP RIMWS RDP LR+SG GN+F+KNL+ SIDNKALYDTFS FG+ILS KV
Sbjct 72 MNFEVIKGQPIRIMWSHRDPGLRKSGMGNIFIKNLENSIDNKALYDTFSTFGSILSSKVV 131
Query 134 VDEFNRSRGYGFVHYETEASARSAI 158
+E SRG+GFVH+ET +A+ AI
Sbjct 132 YNEHG-SRGFGFVHFETHEAAQKAI 155
Score = 81.6 bits (200), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 51/161 (31%), Positives = 90/161 (55%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+ +LH V E L ++F+ G + S++V RDS + +S G+ +VN++ +A++A+D
Sbjct 192 NIYVKNLHANVDEQRLQDLFSQFGNMQSVKVMRDS-NGQSRGFGFVNFEKHEEAQKAVDH 250
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRS------------GAGNVFVKNLDKSIDNKAL 117
+N + GQ + +Q R L+R N++VKNLD SI+++ L
Sbjct 251 MNGKEVSGQLLYVGRAQKRAERQSELKRRFEQMKQERQNRYQGVNLYVKNLDDSINDERL 310
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
+ FS +G I S KV + E + S+G+GFV + + A A+
Sbjct 311 KEVFSTYGVITSAKV-MTESSHSKGFGFVCFSSPEEATKAV 350
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 44/152 (28%), Positives = 84/152 (55%), Gaps = 8/152 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L++ F+ G + S +V + R G+ +V++++ A++A++
Sbjct 100 NIFIKNLENSIDNKALYDTFSTFGSILSSKVVYNEHGSR--GFGFVHFETHEAAQKAINT 157
Query 74 LNFALIRGQPCRI----MWSQRDPSL--RRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ + + +R+ L R G N++VKNL ++D + L D FS FGN+
Sbjct 158 MNGMLLNDRKVFVGHFKSRQKREAELGARALGFTNIYVKNLHANVDEQRLQDLFSQFGNM 217
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
S KV D +SRG+GFV++E A+ A++
Sbjct 218 QSVKVMRDSNGQSRGFGFVNFEKHEEAQKAVD 249
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 44/82 (53%), Gaps = 2/82 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L + + L EVF+ G +TS +V + S S G+ +V + S +A +
Sbjct 291 YQGVNLYVKNLDDSINDERLKEVFSTYGVITSAKVMTE--SSHSKGFGFVCFSSPEEATK 348
Query 70 ALDALNFALIRGQPCRIMWSQR 91
A+ +N ++ +P + +QR
Sbjct 349 AVTEMNGRIVGTKPLYVALAQR 370
> hsa:132430 PABPC4L, DKFZp686J06116; poly(A) binding protein,
cytoplasmic 4-like; K13126 polyadenylate-binding protein
Length=428
Score = 199 bits (507), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 93/151 (61%), Positives = 117/151 (77%), Gaps = 1/151 (0%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y SLY+GDLH +VTE +LF F+ VGPV SIR+CRD V+RRSLGYAYVN+ L DA++
Sbjct 65 YRMASLYVGDLHADVTEDLLFRKFSTVGPVLSIRICRDQVTRRSLGYAYVNFLQLADAQK 124
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
ALD +NF +I+G+ R+MWSQRD LRRSG GNVF+KNLDKSIDNK LY+ FS FG ILS
Sbjct 125 ALDTMNFDIIKGKSIRLMWSQRDAYLRRSGIGNVFIKNLDKSIDNKTLYEHFSAFGKILS 184
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
KV D+ S+GY FVH++ +++A AIE+
Sbjct 185 SKVMSDD-QGSKGYAFVHFQNQSAADRAIEE 214
Score = 88.6 bits (218), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 49/155 (31%), Positives = 90/155 (58%), Gaps = 12/155 (7%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L+E F+A G + S +V D + S GYA+V++Q+ A+RA++
Sbjct 157 NVFIKNLDKSIDNKTLYEHFSAFGKILSSKVMSDD--QGSKGYAFVHFQNQSAADRAIEE 214
Query 74 LNFALIRGQPCRIMWSQ------RDPSLRRSGA--GNVFVKNLDKSIDNKALYDTFSLFG 125
+N L++G C++ + R+ LR + NV++KN +D++ L D FS +G
Sbjct 215 MNGKLLKG--CKVFVGRFKNRKDREAELRSKASEFTNVYIKNFGGDMDDERLKDVFSKYG 272
Query 126 NILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LS KV D +S+G+GFV +++ +A+ A+E+
Sbjct 273 KTLSVKVMTDSSGKSKGFGFVSFDSHEAAKKAVEE 307
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 44/161 (27%), Positives = 84/161 (52%), Gaps = 18/161 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++YI + ++ + L +VF+ G S++V DS S +S G+ +V++ S A++A++
Sbjct 249 NVYIKNFGGDMDDERLKDVFSKYGKTLSVKVMTDS-SGKSKGFGFVSFDSHEAAKKAVEE 307
Query 74 LNFALIRGQ---------------PCRIMWSQ-RDPSLRRSGAGNVFVKNLDKSIDNKAL 117
+N I GQ + M+ Q + +R +++KNLD +ID++ L
Sbjct 308 MNGRDINGQLIFVGRAQKKVERQAELKQMFEQLKRERIRGCQGVKLYIKNLDDTIDDEKL 367
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
+ FS FG+I KV + E +S+G+G + + + A A+
Sbjct 368 RNEFSSFGSISRVKV-MQEEGQSKGFGLICFSSPEDATKAM 407
> xla:495336 hypothetical LOC495336
Length=711
Score = 197 bits (502), Expect = 1e-50, Method: Composition-based stats.
Identities = 87/147 (59%), Positives = 113/147 (76%), Gaps = 0/147 (0%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SLY+GDLHP++ + L F+ +GPV VCRD SR+SLGY YVN++ DAERAL+
Sbjct 3 SLYVGDLHPDINDDQLRMKFSEIGPVAVAHVCRDVTSRKSLGYGYVNFEDPKDAERALEQ 62
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVG 133
+N+ ++ G+P RIMWSQRDPSLR+SG GN+F+KNL K+I+ K LYDTFSLFG ILSCK+
Sbjct 63 MNYEVVMGRPIRIMWSQRDPSLRKSGLGNIFIKNLAKTIEQKELYDTFSLFGRILSCKIA 122
Query 134 VDEFNRSRGYGFVHYETEASARSAIEK 160
+DE S+GYGFVH+E E A+ AI+K
Sbjct 123 MDENGNSKGYGFVHFENEECAKRAIQK 149
Score = 71.2 bits (173), Expect = 1e-12, Method: Composition-based stats.
Identities = 52/162 (32%), Positives = 80/162 (49%), Gaps = 18/162 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+ + PE + L E+F G + S V +DS +S G+ +V Y + AE A+ A
Sbjct 182 NIYVKNFPPETDDEKLKEMFTEFGEIKSACVMKDS-EGKSKGFGFVCYLNPEHAEAAVAA 240
Query 74 LNFALIRGQPCRIMWSQR--------------DPSLRRS---GAGNVFVKNLDKSIDNKA 116
++ I G+ +QR + RRS N++VKNLD ID++
Sbjct 241 MHGKEIGGRSLYASRAQRKEERQEELKLRLEKQKAERRSKYVSNVNLYVKNLDDEIDDER 300
Query 117 LYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
L + FS +G I S KV D NRS+G+GFV + A A+
Sbjct 301 LKEIFSKYGPISSAKVMTDSNNRSKGFGFVCFTNPEQATKAV 342
Score = 63.9 bits (154), Expect = 2e-10, Method: Composition-based stats.
Identities = 42/149 (28%), Positives = 76/149 (51%), Gaps = 5/149 (3%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + + L++ F+ G + S ++ D + S GY +V++++ A+RA+
Sbjct 91 NIFIKNLAKTIEQKELYDTFSLFGRILSCKIAMDE-NGNSKGYGFVHFENEECAKRAIQK 149
Query 74 LNFALIRGQPCR----IMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
+N I G+ I S R R+ N++VKN D++ L + F+ FG I S
Sbjct 150 VNNMSICGKVVYVGNFIPRSDRKSQNRKQKFNNIYVKNFPPETDDEKLKEMFTEFGEIKS 209
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAI 158
V D +S+G+GFV Y A +A+
Sbjct 210 ACVMKDSEGKSKGFGFVCYLNPEHAEAAV 238
> sce:YER165W PAB1; Pab1p; K13126 polyadenylate-binding protein
Length=577
Score = 197 bits (502), Expect = 1e-50, Method: Composition-based stats.
Identities = 85/148 (57%), Positives = 118/148 (79%), Gaps = 0/148 (0%)
Query 12 SPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERAL 71
S SLY+GDL P V+EA L+++F+ +G V+SIRVCRD++++ SLGYAYVN+ +A+
Sbjct 37 SASLYVGDLEPSVSEAHLYDIFSPIGSVSSIRVCRDAITKTSLGYAYVNFNDHEAGRKAI 96
Query 72 DALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCK 131
+ LN+ I+G+ CRIMWSQRDPSLR+ G+GN+F+KNL IDNKALYDTFS+FG+ILS K
Sbjct 97 EQLNYTPIKGRLCRIMWSQRDPSLRKKGSGNIFIKNLHPDIDNKALYDTFSVFGDILSSK 156
Query 132 VGVDEFNRSRGYGFVHYETEASARSAIE 159
+ DE +S+G+GFVH+E E +A+ AI+
Sbjct 157 IATDENGKSKGFGFVHFEEEGAAKEAID 184
Score = 84.0 bits (206), Expect = 2e-16, Method: Composition-based stats.
Identities = 46/154 (29%), Positives = 86/154 (55%), Gaps = 7/154 (4%)
Query 12 SPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERAL 71
S +++I +LHP++ L++ F+ G + S ++ D + +S G+ +V+++ G A+ A+
Sbjct 125 SGNIFIKNLHPDIDNKALYDTFSVFGDILSSKIATDE-NGKSKGFGFVHFEEEGAAKEAI 183
Query 72 DALNFALIRGQPCRIM----WSQRDPSLRRSGA--GNVFVKNLDKSIDNKALYDTFSLFG 125
DALN L+ GQ + +RD L + A N++VKN++ ++ + F+ FG
Sbjct 184 DALNGMLLNGQEIYVAPHLSRKERDSQLEETKAHYTNLYVKNINSETTDEQFQELFAKFG 243
Query 126 NILSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
I+S + D + +G+GFV+YE A A+E
Sbjct 244 PIVSASLEKDADGKLKGFGFVNYEKHEDAVKAVE 277
Score = 80.9 bits (198), Expect = 2e-15, Method: Composition-based stats.
Identities = 49/161 (30%), Positives = 85/161 (52%), Gaps = 17/161 (10%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+ +++ E T+ E+F GP+ S + +D+ + G+ +VNY+ DA +A++A
Sbjct 220 NLYVKNINSETTDEQFQELFAKFGPIVSASLEKDADGKLK-GFGFVNYEKHEDAVKAVEA 278
Query 74 LNFALIRGQPCRIMWSQ----------------RDPSLRRSGAGNVFVKNLDKSIDNKAL 117
LN + + G+ + +Q R + + N+FVKNLD S+D++ L
Sbjct 279 LNDSELNGEKLYVGRAQKKNERMHVLKKQYEAYRLEKMAKYQGVNLFVKNLDDSVDDEKL 338
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
+ F+ +G I S KV E +S+G+GFV + T A AI
Sbjct 339 EEEFAPYGTITSAKVMRTENGKSKGFGFVCFSTPEEATKAI 379
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 98 SGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFNR-SRGYGFVHYETEASARS 156
+ + +++V +L+ S+ LYD FS G++ S +V D + S GY +V++ + R
Sbjct 35 NSSASLYVGDLEPSVSEAHLYDIFSPIGSVSSIRVCRDAITKTSLGYAYVNFNDHEAGRK 94
Query 157 AIEK 160
AIE+
Sbjct 95 AIEQ 98
Score = 39.3 bits (90), Expect = 0.006, Method: Composition-based stats.
Identities = 25/98 (25%), Positives = 49/98 (50%), Gaps = 1/98 (1%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +L++ +L V + L E F G +TS +V R + + +S G+ +V + + +A +
Sbjct 319 YQGVNLFVKNLDDSVDDEKLEEEFAPYGTITSAKVMR-TENGKSKGFGFVCFSTPEEATK 377
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKN 107
A+ N ++ G+P + +QR R A + +N
Sbjct 378 AITEKNQQIVAGKPLYVAIAQRKDVRRSQLAQQIQARN 415
> mmu:241989 Pabpc4l, C330050A14Rik, EG241989; poly(A) binding
protein, cytoplasmic 4-like; K13126 polyadenylate-binding protein
Length=370
Score = 194 bits (494), Expect = 7e-50, Method: Compositional matrix adjust.
Identities = 92/154 (59%), Positives = 115/154 (74%), Gaps = 1/154 (0%)
Query 7 HPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGD 66
Y + SLY+GDLH +VTE MLF F+ VGPV SIR+CRD +S+RSLGYAYVN+ + D
Sbjct 4 EAKYRAASLYVGDLHEDVTEDMLFRKFSTVGPVLSIRICRDLISQRSLGYAYVNFLQVND 63
Query 67 AERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGN 126
A++AL +NF +I+G+ R+MWSQRD LRRSG GNVF+KNLDKSIDNK LY+ FS FG
Sbjct 64 AQKALVTMNFDVIKGKSIRLMWSQRDACLRRSGVGNVFIKNLDKSIDNKTLYEHFSPFGT 123
Query 127 ILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
I+S KV D S+GYGFVHY+ +A AIE+
Sbjct 124 IMSSKVMTDG-EGSKGYGFVHYQDRRAADRAIEE 156
Score = 78.6 bits (192), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 46/152 (30%), Positives = 81/152 (53%), Gaps = 8/152 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L+E F+ G + S +V D S GY +V+YQ A+RA++
Sbjct 99 NVFIKNLDKSIDNKTLYEHFSPFGTIMSSKVMTDGEG--SKGYGFVHYQDRRAADRAIEE 156
Query 74 LNFALIRGQPCRIMW----SQRDPSLRRSGA--GNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+R + R+ LR NV++KN +D++ L + FS +G
Sbjct 157 MNGKLLRESTLFVARFKSRKDREAELRDKPTEFTNVYIKNFGDDVDDEKLREVFSKYGQT 216
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
LS KV D +S+G+GFV +++ +A++A+E
Sbjct 217 LSVKVMKDATGKSKGFGFVSFDSHEAAKNAVE 248
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 45/157 (28%), Positives = 83/157 (52%), Gaps = 19/157 (12%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
PT + ++YI + +V + L EVF+ G S++V +D+ + S G+ +V++ S A
Sbjct 186 PTEFT-NVYIKNFGDDVDDEKLREVFSKYGQTLSVKVMKDATGK-SKGFGFVSFDSHEAA 243
Query 68 ERALDALNFALIRGQPCRI---------------MWSQ-RDPSLRRSGAGNVFVKNLDKS 111
+ A++ +N I GQ + M+ Q + +R A +++KNLD +
Sbjct 244 KNAVEDMNGQDINGQTIFVGRAQKKVERQAELKEMFEQMKKERIRARQAAKLYIKNLDDT 303
Query 112 IDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHY 148
ID++ L FS+FG+I KV + E +S+G+G + +
Sbjct 304 IDDETLRKEFSVFGSICRVKV-MQEAGQSKGFGLICF 339
> ath:AT4G34110 PAB2; PAB2 (POLY(A) BINDING 2); RNA binding /
translation initiation factor; K13126 polyadenylate-binding
protein
Length=629
Score = 194 bits (494), Expect = 8e-50, Method: Compositional matrix adjust.
Identities = 89/147 (60%), Positives = 115/147 (78%), Gaps = 0/147 (0%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SLY+GDL VT++ LF+ F +G V ++RVCRD V+RRSLGY YVN+ + DA RA+
Sbjct 37 SLYVGDLDFNVTDSQLFDAFGQMGTVVTVRVCRDLVTRRSLGYGYVNFTNPQDAARAIQE 96
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVG 133
LN+ + G+P R+M+S RDPS+RRSGAGN+F+KNLD+SID+KAL+DTFS FGNI+SCKV
Sbjct 97 LNYIPLYGKPIRVMYSHRDPSVRRSGAGNIFIKNLDESIDHKALHDTFSSFGNIVSCKVA 156
Query 134 VDEFNRSRGYGFVHYETEASARSAIEK 160
VD +S+GYGFV Y E SA+ AIEK
Sbjct 157 VDSSGQSKGYGFVQYANEESAQKAIEK 183
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 45/150 (30%), Positives = 80/150 (53%), Gaps = 5/150 (3%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L + F++ G + S +V DS S +S GY +V Y + A++A++
Sbjct 125 NIFIKNLDESIDHKALHDTFSSFGNIVSCKVAVDS-SGQSKGYGFVQYANEESAQKAIEK 183
Query 74 LNFALIRGQPCRI----MWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
LN L+ + + +RD + ++ NV+VKNL +S + L + F +G I S
Sbjct 184 LNGMLLNDKQVYVGPFLRRQERDSTANKTKFTNVYVKNLAESTTDDDLKNAFGEYGKITS 243
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAIE 159
V D +S+G+GFV++E A A+E
Sbjct 244 AVVMKDGEGKSKGFGFVNFENADDAARAVE 273
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 49/163 (30%), Positives = 81/163 (49%), Gaps = 17/163 (10%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+ +L T+ L F G +TS V +D +S G+ +VN+++ DA RA+++
Sbjct 216 NVYVKNLAESTTDDDLKNAFGEYGKITSAVVMKDG-EGKSKGFGFVNFENADDAARAVES 274
Query 74 LNFALIRGQPCRI----MWSQRDPSLR------------RSGAGNVFVKNLDKSIDNKAL 117
LN + + S+R+ LR + + N++VKNLD SI ++ L
Sbjct 275 LNGHKFDDKEWYVGRAQKKSERETELRVRYEQNLKEAADKFQSSNLYVKNLDPSISDEKL 334
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
+ FS FG + S KV D S+G GFV + T A A+ +
Sbjct 335 KEIFSPFGTVTSSKVMRDPNGTSKGSGFVAFATPEEATEAMSQ 377
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 47/87 (54%), Gaps = 1/87 (1%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
+ S +LY+ +L P +++ L E+F+ G VTS +V RD + S G +V + + +A
Sbjct 315 FQSSNLYVKNLDPSISDEKLKEIFSPFGTVTSSKVMRDP-NGTSKGSGFVAFATPEEATE 373
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLR 96
A+ L+ +I +P + +QR R
Sbjct 374 AMSQLSGKMIESKPLYVAIAQRKEDRR 400
> ath:AT1G71770 PAB5; PAB5 (POLY(A)-BINDING PROTEIN 5); RNA binding
/ poly(A) binding / translation initiation factor
Length=668
Score = 194 bits (493), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 92/152 (60%), Positives = 118/152 (77%), Gaps = 1/152 (0%)
Query 9 TYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAE 68
T+ + SLY+GDL P V E+ L ++FN V PV ++RVCRD ++ RSLGYAYVN+ + DA
Sbjct 41 THPNSSLYVGDLDPSVNESHLLDLFNQVAPVHNLRVCRD-LTHRSLGYAYVNFANPEDAS 99
Query 69 RALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNIL 128
RA+++LN+A IR +P RIM S RDPS R SG GNVF+KNLD SIDNKALY+TFS FG IL
Sbjct 100 RAMESLNYAPIRDRPIRIMLSNRDPSTRLSGKGNVFIKNLDASIDNKALYETFSSFGTIL 159
Query 129 SCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
SCKV +D RS+GYGFV +E E +A++AI+K
Sbjct 160 SCKVAMDVVGRSKGYGFVQFEKEETAQAAIDK 191
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 50/153 (32%), Positives = 80/153 (52%), Gaps = 7/153 (4%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L+E F++ G + S +V D V R S GY +V ++ A+ A+D
Sbjct 133 NVFIKNLDASIDNKALYETFSSFGTILSCKVAMDVVGR-SKGYGFVQFEKEETAQAAIDK 191
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAG------NVFVKNLDKSIDNKALYDTFSLFGNI 127
LN L+ + + R RS +G NV+VKNL K I + L TF +G+I
Sbjct 192 LNGMLLNDKQVFVGHFVRRQDRARSESGAVPSFTNVYVKNLPKEITDDELKKTFGKYGDI 251
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
S V D+ SR +GFV++ + +A A+EK
Sbjct 252 SSAVVMKDQSGNSRSFGFVNFVSPEAAAVAVEK 284
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 54/178 (30%), Positives = 89/178 (50%), Gaps = 25/178 (14%)
Query 1 DAGAGPHPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVN 60
++GA P T V Y+ +L E+T+ L + F G ++S V +D S S + +VN
Sbjct 217 ESGAVPSFTNV----YVKNLPKEITDDELKKTFGKYGDISSAVVMKDQ-SGNSRSFGFVN 271
Query 61 YQSLGDAERALDALNF------ALIRGQPCRIMWSQRDPSLRRS------------GAGN 102
+ S A A++ +N L G+ + S R+ LRR N
Sbjct 272 FVSPEAAAVAVEKMNGISLGEDVLYVGRAQK--KSDREEELRRKFEQERISRFEKLQGSN 329
Query 103 VFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
+++KNLD S++++ L + FS +GN+ SCKV ++ SRG+GFV Y A A+++
Sbjct 330 LYLKNLDDSVNDEKLKEMFSEYGNVTSCKVMMNSQGLSRGFGFVAYSNPEEALLAMKE 387
> ath:AT1G49760 PAB8; PAB8 (POLY(A) BINDING PROTEIN 8); RNA binding
/ translation initiation factor; K13126 polyadenylate-binding
protein
Length=671
Score = 193 bits (491), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 89/147 (60%), Positives = 113/147 (76%), Gaps = 0/147 (0%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SLY+GDL VT++ LFE F G V S+RVCRD +RRSLGY YVNY + DA RAL+
Sbjct 46 SLYVGDLDATVTDSQLFEAFTQAGQVVSVRVCRDMTTRRSLGYGYVNYATPQDASRALNE 105
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVG 133
LNF + G+ R+M+S RDPSLR+SG GN+F+KNLDKSID+KAL++TFS FG ILSCKV
Sbjct 106 LNFMALNGRAIRVMYSVRDPSLRKSGVGNIFIKNLDKSIDHKALHETFSAFGPILSCKVA 165
Query 134 VDEFNRSRGYGFVHYETEASARSAIEK 160
VD +S+GYGFV Y+T+ +A+ AI+K
Sbjct 166 VDPSGQSKGYGFVQYDTDEAAQGAIDK 192
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 50/150 (33%), Positives = 80/150 (53%), Gaps = 5/150 (3%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + L E F+A GP+ S +V D S +S GY +V Y + A+ A+D
Sbjct 134 NIFIKNLDKSIDHKALHETFSAFGPILSCKVAVDP-SGQSKGYGFVQYDTDEAAQGAIDK 192
Query 74 LNFALIRGQPCRI----MWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
LN L+ + + QRDPS + NV+VKNL +S+ ++ L F FG S
Sbjct 193 LNGMLLNDKQVYVGPFVHKLQRDPSGEKVKFTNVYVKNLSESLSDEELNKVFGEFGVTTS 252
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAIE 159
C + D +S+G+GFV++E A A++
Sbjct 253 CVIMRDGEGKSKGFGFVNFENSDDAARAVD 282
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 53/161 (32%), Positives = 84/161 (52%), Gaps = 17/161 (10%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+ +L +++ L +VF G TS + RD + S G+ +VN+++ DA RA+DA
Sbjct 225 NVYVKNLSESLSDEELNKVFGEFGVTTSCVIMRDGEGK-SKGFGFVNFENSDDAARAVDA 283
Query 74 LNFALIRGQPCRIMWSQR------------DPSLR----RSGAGNVFVKNLDKSIDNKAL 117
LN + + +Q+ + SL+ +S N++VKNLD+S+ + L
Sbjct 284 LNGKTFDDKEWFVGKAQKKSERETELKQKFEQSLKEAADKSQGSNLYVKNLDESVTDDKL 343
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
+ F+ FG I SCKV D SRG GFV + T A AI
Sbjct 344 REHFAPFGTITSCKVMRDPSGVSRGSGFVAFSTPEEATRAI 384
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 1/78 (1%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+ +L VT+ L E F G +TS +V RD S S G +V + + +A RA+
Sbjct 328 NLYVKNLDESVTDDKLREHFAPFGTITSCKVMRDP-SGVSRGSGFVAFSTPEEATRAITE 386
Query 74 LNFALIRGQPCRIMWSQR 91
+N +I +P + +QR
Sbjct 387 MNGKMIVTKPLYVALAQR 404
> ath:AT1G22760 PAB3; PAB3 (POLY(A) BINDING PROTEIN 3); RNA binding
/ translation initiation factor
Length=660
Score = 188 bits (477), Expect = 7e-48, Method: Compositional matrix adjust.
Identities = 91/152 (59%), Positives = 116/152 (76%), Gaps = 1/152 (0%)
Query 9 TYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAE 68
T+ + SLY GDL P+VTEA LF++F V V S+RVCRD +RRSLGYAY+N+ + DA
Sbjct 45 THPNSSLYAGDLDPKVTEAHLFDLFKHVANVVSVRVCRDQ-NRRSLGYAYINFSNPNDAY 103
Query 69 RALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNIL 128
RA++ALN+ + +P RIM S RDPS R SG GN+F+KNLD SIDNKAL++TFS FG IL
Sbjct 104 RAMEALNYTPLFDRPIRIMLSNRDPSTRLSGKGNIFIKNLDASIDNKALFETFSSFGTIL 163
Query 129 SCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
SCKV +D RS+GYGFV +E E SA++AI+K
Sbjct 164 SCKVAMDVTGRSKGYGFVQFEKEESAQAAIDK 195
Score = 82.0 bits (201), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 54/153 (35%), Positives = 78/153 (50%), Gaps = 7/153 (4%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + LFE F++ G + S +V D V+ RS GY +V ++ A+ A+D
Sbjct 137 NIFIKNLDASIDNKALFETFSSFGTILSCKVAMD-VTGRSKGYGFVQFEKEESAQAAIDK 195
Query 74 LNFALIR------GQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
LN L+ G R RD + NV+VKNL K I L TF FG I
Sbjct 196 LNGMLMNDKQVFVGHFIRRQERARDENTPTPRFTNVYVKNLPKEIGEDELRKTFGKFGVI 255
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
S V D+ SR +GFV++E +A SA+EK
Sbjct 256 SSAVVMRDQSGNSRCFGFVNFECTEAAASAVEK 288
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 52/171 (30%), Positives = 85/171 (49%), Gaps = 21/171 (12%)
Query 8 PTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDA 67
PT ++Y+ +L E+ E L + F G ++S V RD S S + +VN++ A
Sbjct 224 PTPRFTNVYVKNLPKEIGEDELRKTFGKFGVISSAVVMRDQ-SGNSRCFGFVNFECTEAA 282
Query 68 ERALDALNF------ALIRGQPCRIMWSQRDPSLRR------------SGAGNVFVKNLD 109
A++ +N L G+ + S+R+ LRR S N+++KNLD
Sbjct 283 ASAVEKMNGISLGDDVLYVGRAQK--KSEREEELRRKFEQERINRFEKSQGANLYLKNLD 340
Query 110 KSIDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
S+D++ L + FS +GN+ S KV ++ SRG+GFV Y A A+ +
Sbjct 341 DSVDDEKLKEMFSEYGNVTSSKVMLNPQGMSRGFGFVAYSNPEEALRALSE 391
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 1/78 (1%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+ +L V + L E+F+ G VTS +V + S G+ +V Y + +A RAL
Sbjct 333 NLYLKNLDDSVDDEKLKEMFSEYGNVTSSKVMLNPQGM-SRGFGFVAYSNPEEALRALSE 391
Query 74 LNFALIRGQPCRIMWSQR 91
+N +I +P I +QR
Sbjct 392 MNGKMIGRKPLYIALAQR 409
> hsa:140886 PABPC5, FLJ51677, PABP5; poly(A) binding protein,
cytoplasmic 5; K13126 polyadenylate-binding protein
Length=382
Score = 171 bits (433), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 81/142 (57%), Positives = 106/142 (74%), Gaps = 1/142 (0%)
Query 17 IGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDALNF 76
+GDL P+VTE ML++ F GP+ R+CRD V+R LGY YVN++ DAE AL+ +NF
Sbjct 22 VGDLDPDVTEDMLYKKFRPAGPLRFTRICRDPVTRSPLGYGYVNFRFPADAEWALNTMNF 81
Query 77 ALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDE 136
LI G+P R+MWSQ D LR+SG GN+F+KNLDKSIDN+AL+ FS FGNILSCKV D+
Sbjct 82 DLINGKPFRLMWSQPDDRLRKSGVGNIFIKNLDKSIDNRALFYLFSAFGNILSCKVVCDD 141
Query 137 FNRSRGYGFVHYETEASARSAI 158
N S+GY +VH+++ A+A AI
Sbjct 142 -NGSKGYAYVHFDSLAAANRAI 162
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 51/153 (33%), Positives = 83/153 (54%), Gaps = 11/153 (7%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRV-CRDSVSRRSLGYAYVNYQSLGDAERALD 72
+++I +L + LF +F+A G + S +V C D+ S+ GYAYV++ SL A RA+
Sbjct 107 NIFIKNLDKSIDNRALFYLFSAFGNILSCKVVCDDNGSK---GYAYVHFDSLAAANRAIW 163
Query 73 ALNFALIRGQPCRI----MWSQRDPSLR---RSGAGNVFVKNLDKSIDNKALYDTFSLFG 125
+N + + + +R +R R+ NVFVKN+ ID++ L + F +G
Sbjct 164 HMNGVRLNNRQVYVGRFKFPEERAAEVRTRDRATFTNVFVKNIGDDIDDEKLKELFCEYG 223
Query 126 NILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
S KV D +S+G+GFV YET +A+ A+
Sbjct 224 PTESVKVIRDASGKSKGFGFVRYETHEAAQKAV 256
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 40/163 (24%), Positives = 85/163 (52%), Gaps = 18/163 (11%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++++ ++ ++ + L E+F GP S++V RD+ S +S G+ +V Y++ A++A+
Sbjct 200 NVFVKNIGDDIDDEKLKELFCEYGPTESVKVIRDA-SGKSKGFGFVRYETHEAAQKAVLD 258
Query 74 LNFALIRGQPCRIMWSQRD----------------PSLRRSGAGNVFVKNLDKSIDNKAL 117
L+ I G+ + +Q+ R +++KNLD++I+++ L
Sbjct 259 LHGKSIDGKVLYVGRAQKKIERLAELRRRFERLRLKEKSRPPGVPIYIKNLDETINDEKL 318
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
+ FS FG+I KV + E + +G+G V + + A A+++
Sbjct 319 KEEFSSFGSISRAKVMM-EVGQGKGFGVVCFSSFEEATKAVDE 360
> mmu:93728 Pabpc5, C820015E17, MGC130340, MGC130341; poly(A)
binding protein, cytoplasmic 5; K13126 polyadenylate-binding
protein
Length=381
Score = 170 bits (430), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 80/142 (56%), Positives = 106/142 (74%), Gaps = 1/142 (0%)
Query 17 IGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDALNF 76
+GDL P+VTE ML++ F GP+ R+CRD V+R LGY YVN++ DAE AL+ +NF
Sbjct 21 VGDLDPDVTEDMLYKKFRPAGPLRFTRICRDPVTRSPLGYGYVNFRFPADAEWALNTMNF 80
Query 77 ALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDE 136
LI G+P R+MWSQ D LR+SG GN+F+KNLDK+IDN+AL+ FS FGNILSCKV D+
Sbjct 81 DLINGKPFRLMWSQPDDRLRKSGVGNIFIKNLDKTIDNRALFYLFSAFGNILSCKVVCDD 140
Query 137 FNRSRGYGFVHYETEASARSAI 158
N S+GY +VH+++ A+A AI
Sbjct 141 -NGSKGYAYVHFDSLAAANRAI 161
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 52/153 (33%), Positives = 82/153 (53%), Gaps = 11/153 (7%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRV-CRDSVSRRSLGYAYVNYQSLGDAERALD 72
+++I +L + LF +F+A G + S +V C D+ S+ GYAYV++ SL A RA+
Sbjct 106 NIFIKNLDKTIDNRALFYLFSAFGNILSCKVVCDDNGSK---GYAYVHFDSLAAANRAIW 162
Query 73 ALNFALIRGQPCRI----MWSQRDPSLR---RSGAGNVFVKNLDKSIDNKALYDTFSLFG 125
+N + + + +R +R R+ NVFVKN ID++ L FS +G
Sbjct 163 HMNGVRLNNRQVYVGRFKFPEERAAEVRTRERATFTNVFVKNFGDDIDDEKLNKLFSEYG 222
Query 126 NILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
S KV D +S+G+GFV YET +A+ A+
Sbjct 223 PTESVKVIRDATGKSKGFGFVRYETHEAAQKAV 255
Score = 58.5 bits (140), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 42/164 (25%), Positives = 88/164 (53%), Gaps = 20/164 (12%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++++ + ++ + L ++F+ GP S++V RD+ + S G+ +V Y++ A++A+
Sbjct 199 NVFVKNFGDDIDDEKLNKLFSEYGPTESVKVIRDATGK-SKGFGFVRYETHEAAQKAVLE 257
Query 74 LNFALIRGQPCRIMWSQRD----PSLRR-------------SGAGNVFVKNLDKSIDNKA 116
L+ I G+ + +Q+ LRR SG +++KNLD++I+++
Sbjct 258 LHGKSIDGKVLCVGRAQKKIERLAELRRRFERLKLKEKNRPSGVP-IYIKNLDETINDEK 316
Query 117 LYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
L + FS FG+I KV + E + +G+G V + + A A+++
Sbjct 317 LKEEFSSFGSISRAKVMM-EVGQGKGFGVVCFSSFEEACKAVDE 359
> ath:AT2G36660 PAB7; PAB7 (POLY(A) BINDING PROTEIN 7); RNA binding
/ translation initiation factor
Length=609
Score = 162 bits (409), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 76/149 (51%), Positives = 102/149 (68%), Gaps = 0/149 (0%)
Query 11 VSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERA 70
V+ SLY+GDLHP VTE +L++ F +TS+R+C+D+ S RSL Y Y N+ S DA A
Sbjct 22 VTASLYVGDLHPSVTEGILYDAFAEFKSLTSVRLCKDASSGRSLCYGYANFLSRQDANLA 81
Query 71 LDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSC 130
++ N +L+ G+ R+MWS R P RR+G GNVFVKNL +S+ N L D F FGNI+SC
Sbjct 82 IEKKNNSLLNGKMIRVMWSVRAPDARRNGVGNVFVKNLPESVTNAVLQDMFKKFGNIVSC 141
Query 131 KVGVDEFNRSRGYGFVHYETEASARSAIE 159
KV E +SRGYGFV +E E +A +AI+
Sbjct 142 KVATLEDGKSRGYGFVQFEQEDAAHAAIQ 170
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 50/171 (29%), Positives = 85/171 (49%), Gaps = 21/171 (12%)
Query 6 PHPTYVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLG 65
P Y + LY+ +L +V+E +L E F G + S+ + +D +R GYA+VN+ +
Sbjct 196 PEEKYTN--LYMKNLDADVSEDLLREKFAEFGKIVSLAIAKDE-NRLCRGYAFVNFDNPE 252
Query 66 DAERALDALNFALIRGQPCRIM-----WSQRDPSLRR------------SGAGNVFVKNL 108
DA RA + +N G C + ++R+ LR + N++VKN+
Sbjct 253 DARRAAETVNGTKF-GSKCLYVGRAQKKAEREQLLREQFKEKHEEQKMIAKVSNIYVKNV 311
Query 109 DKSIDNKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
+ ++ + L FS G I S K+ DE +S+G+GFV + T A A++
Sbjct 312 NVAVTEEELRKHFSQCGTITSTKLMCDEKGKSKGFGFVCFSTPEEAIDAVK 362
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 72/150 (48%), Gaps = 7/150 (4%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVC--RDSVSRRSLGYAYVNYQSLGDAERAL 71
++++ +L VT A+L ++F G + S +V D SR GY +V ++ A A+
Sbjct 113 NVFVKNLPESVTNAVLQDMFKKFGNIVSCKVATLEDGKSR---GYGFVQFEQEDAAHAAI 169
Query 72 DALNFALIRGQPCRI--MWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
LN ++ + + + D N+++KNLD + L + F+ FG I+S
Sbjct 170 QTLNSTIVADKEIYVGKFMKKTDRVKPEEKYTNLYMKNLDADVSEDLLREKFAEFGKIVS 229
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAIE 159
+ DE RGY FV+++ AR A E
Sbjct 230 LAIAKDENRLCRGYAFVNFDNPEDARRAAE 259
Score = 32.0 bits (71), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 34/61 (55%), Gaps = 1/61 (1%)
Query 101 GNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFN-RSRGYGFVHYETEASARSAIE 159
+++V +L S+ LYD F+ F ++ S ++ D + RS YG+ ++ + A AIE
Sbjct 24 ASLYVGDLHPSVTEGILYDAFAEFKSLTSVRLCKDASSGRSLCYGYANFLSRQDANLAIE 83
Query 160 K 160
K
Sbjct 84 K 84
> ath:AT2G23350 PAB4; PAB4 (POLY(A) BINDING PROTEIN 4); RNA binding
/ translation initiation factor
Length=662
Score = 157 bits (396), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 81/147 (55%), Positives = 107/147 (72%), Gaps = 0/147 (0%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SLY+GDL VT++ L++ F V V S+RVCRD+ + SLGY YVNY + DAE+A+
Sbjct 47 SLYVGDLDFNVTDSQLYDYFTEVCQVVSVRVCRDAATNTSLGYGYVNYSNTDDAEKAMQK 106
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVG 133
LN++ + G+ RI +S RD S RRSG GN+FVKNLDKS+DNK L++ FS G I+SCKV
Sbjct 107 LNYSYLNGKMIRITYSSRDSSARRSGVGNLFVKNLDKSVDNKTLHEAFSGCGTIVSCKVA 166
Query 134 VDEFNRSRGYGFVHYETEASARSAIEK 160
D +SRGYGFV ++TE SA++AIEK
Sbjct 167 TDHMGQSRGYGFVQFDTEDSAKNAIEK 193
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 76/150 (50%), Gaps = 5/150 (3%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+L++ +L V L E F+ G + S +V D + + S GY +V + + A+ A++
Sbjct 135 NLFVKNLDKSVDNKTLHEAFSGCGTIVSCKVATDHMGQ-SRGYGFVQFDTEDSAKNAIEK 193
Query 74 LNFALIRGQPCRI----MWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
LN ++ + + +R+ + + NV+VKNL ++ + L TF +G+I S
Sbjct 194 LNGKVLNDKQIFVGPFLRKEERESAADKMKFTNVYVKNLSEATTDDELKTTFGQYGSISS 253
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAIE 159
V D +SR +GFV++E A A+E
Sbjct 254 AVVMRDGDGKSRCFGFVNFENPEDAARAVE 283
Score = 65.1 bits (157), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 49/157 (31%), Positives = 77/157 (49%), Gaps = 17/157 (10%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+ +L T+ L F G ++S V RD +S + +VN+++ DA RA++A
Sbjct 226 NVYVKNLSEATTDDELKTTFGQYGSISSAVVMRDG-DGKSRCFGFVNFENPEDAARAVEA 284
Query 74 LNFALIRGQPCRI----MWSQRDPSLRR------SGAGNVF------VKNLDKSIDNKAL 117
LN + + S+R+ L R S GN F VKNLD ++ ++ L
Sbjct 285 LNGKKFDDKEWYVGKAQKKSERELELSRRYEQGSSDGGNKFDGLNLYVKNLDDTVTDEKL 344
Query 118 YDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASA 154
+ F+ FG I SCKV D S+G GFV + + A
Sbjct 345 RELFAEFGTITSCKVMRDPSGTSKGSGFVAFSAASEA 381
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 45/83 (54%), Gaps = 1/83 (1%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+ +L VT+ L E+F G +TS +V RD S S G +V + + +A R L+
Sbjct 329 NLYVKNLDDTVTDEKLRELFAEFGTITSCKVMRDP-SGTSKGSGFVAFSAASEASRVLNE 387
Query 74 LNFALIRGQPCRIMWSQRDPSLR 96
+N ++ G+P + +QR R
Sbjct 388 MNGKMVGGKPLYVALAQRKEERR 410
> cel:Y106G6H.2 pab-1; PolyA Binding protein family member (pab-1);
K13126 polyadenylate-binding protein
Length=583
Score = 152 bits (385), Expect = 3e-37, Method: Composition-based stats.
Identities = 65/93 (69%), Positives = 82/93 (88%), Gaps = 0/93 (0%)
Query 68 ERALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNI 127
ERA+D +NF + G+P RIMWSQRDP++RRSGAGN+F+KNLDK IDNK++YDTFSLFGNI
Sbjct 24 ERAMDTMNFEALHGKPMRIMWSQRDPAMRRSGAGNIFIKNLDKVIDNKSIYDTFSLFGNI 83
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
LSCKV +DE S+GYGFVH+ETE +A++AI+K
Sbjct 84 LSCKVAIDEDGFSKGYGFVHFETEEAAQNAIQK 116
Score = 72.8 bits (177), Expect = 5e-13, Method: Composition-based stats.
Identities = 50/163 (30%), Positives = 82/163 (50%), Gaps = 20/163 (12%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+ + + L ++F G +TS V +V +S G+ +V + + +AE A+ A
Sbjct 151 NVYVKNFGDHYNKETLEKLFAKFGNITSCEVM--TVEGKSKGFGFVAFANPEEAETAVQA 208
Query 74 LNFALIRG-----QPCRIMW-SQRDPSLRRSGAG------------NVFVKNLDKSIDNK 115
L+ + I G CR S+R L++ N++VKNLD+++D+
Sbjct 209 LHDSTIEGTDLKLHVCRAQKKSERHAELKKKHEQHKAERMQKYQGVNLYVKNLDETVDDD 268
Query 116 ALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAI 158
L F +GNI S KV DE RS+G+GFV +E A SA+
Sbjct 269 GLKKQFESYGNITSAKVMTDENGRSKGFGFVCFEKPEEATSAV 311
Score = 61.2 bits (147), Expect = 2e-09, Method: Composition-based stats.
Identities = 41/152 (26%), Positives = 78/152 (51%), Gaps = 8/152 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+++I +L + +++ F+ G + S +V D S GY +V++++ A+ A+
Sbjct 58 NIFIKNLDKVIDNKSIYDTFSLFGNILSCKVAIDE-DGFSKGYGFVHFETEEAAQNAIQK 116
Query 74 LNFALIRGQPCRI----MWSQRDPSLRRSGA--GNVFVKNLDKSIDNKALYDTFSLFGNI 127
+N L+ G+ + +QR+ L + NV+VKN + + L F+ FGNI
Sbjct 117 VNGMLLAGKKVFVGKFQPRAQRNRELGETAKQFTNVYVKNFGDHYNKETLEKLFAKFGNI 176
Query 128 LSCKVGVDEFNRSRGYGFVHYETEASARSAIE 159
SC+V E +S+G+GFV + A +A++
Sbjct 177 TSCEVMTVE-GKSKGFGFVAFANPEEAETAVQ 207
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 25/99 (25%), Positives = 51/99 (51%), Gaps = 2/99 (2%)
Query 10 YVSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAER 69
Y +LY+ +L V + L + F + G +TS +V D + RS G+ +V ++ +A
Sbjct 251 YQGVNLYVKNLDETVDDDGLKKQFESYGNITSAKVMTDE-NGRSKGFGFVCFEKPEEATS 309
Query 70 ALDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNL 108
A+ +N ++ +P + +QR RR+ + +++ L
Sbjct 310 AVTEMNSKMVCSKPLYVAIAQRKED-RRAQLASQYMQRL 347
> ath:AT3G16380 PAB6; PAB6 (POLY(A) BINDING PROTEIN 6); RNA binding
/ translation initiation factor
Length=537
Score = 137 bits (346), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 72/147 (48%), Positives = 101/147 (68%), Gaps = 4/147 (2%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SLY+GDL P+VTE L + F+ PV S+ +CR+SV+ +S+ YAY+N+ S A A+
Sbjct 22 SLYVGDLSPDVTEKDLIDKFSLNVPVVSVHLCRNSVTGKSMCYAYINFDSPFSASNAMTR 81
Query 74 LNFALIRGQPCRIMWSQRDPSLR---RSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSC 130
LN + ++G+ RIMWSQRD + R R+G N++VKNLD SI + L F FG+ILSC
Sbjct 82 LNHSDLKGKAMRIMWSQRDLAYRRRTRTGFANLYVKNLDSSITSSCLERMFCPFGSILSC 141
Query 131 KVGVDEFNRSRGYGFVHYETEASARSA 157
KV V+E +S+G+GFV ++TE SA SA
Sbjct 142 KV-VEENGQSKGFGFVQFDTEQSAVSA 167
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 50/164 (30%), Positives = 88/164 (53%), Gaps = 22/164 (13%)
Query 12 SPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERAL 71
S ++Y+ +L VT+ L +F+ G V+S+ V RD + R S G+ +VN+ + +A++A+
Sbjct 201 STNVYVKNLIETVTDDCLHTLFSQYGTVSSVVVMRDGMGR-SRGFGFVNFCNPENAKKAM 259
Query 72 DAL------NFALIRGQPC-----RIMWSQR-------DPSLRRSGAGNVFVKNLDKSID 113
++L + L G+ R M Q+ P++R S N++VKNL +S++
Sbjct 260 ESLCGLQLGSKKLFVGKALKKDERREMLKQKFSDNFIAKPNMRWS---NLYVKNLSESMN 316
Query 114 NKALYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSA 157
L + F +G I+S KV E RS+G+GFV + ++ A
Sbjct 317 ETRLREIFGCYGQIVSAKVMCHENGRSKGFGFVCFSNCEESKQA 360
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 81/150 (54%), Gaps = 6/150 (4%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+ +L +T + L +F G + S +V ++ +S G+ +V + + A A A
Sbjct 113 NLYVKNLDSSITSSCLERMFCPFGSILSCKVVEEN--GQSKGFGFVQFDTEQSAVSARSA 170
Query 74 LNFALIRGQPCRI-MWSQRDPSLRRSG---AGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
L+ +++ G+ + + +D +G + NV+VKNL +++ + L+ FS +G + S
Sbjct 171 LHGSMVYGKKLFVAKFINKDERAAMAGNQDSTNVYVKNLIETVTDDCLHTLFSQYGTVSS 230
Query 130 CKVGVDEFNRSRGYGFVHYETEASARSAIE 159
V D RSRG+GFV++ +A+ A+E
Sbjct 231 VVVMRDGMGRSRGFGFVNFCNPENAKKAME 260
> tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12831
splicing factor 3B subunit 4
Length=576
Score = 105 bits (261), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 60/149 (40%), Positives = 94/149 (63%), Gaps = 8/149 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LYIG+L +V + +L+E+F GPV ++ V RD ++ GY +V +++ DA+ AL
Sbjct 30 TLYIGNLDSQVDDDLLWELFVQCGPVRTVSVPRDKLTGNHQGYGFVEFRNEVDADYALKL 89
Query 74 LNFALIRGQPCRIMWSQRDPSLRRS---GAGNVFVKNLDKSIDNKALYDTFSLFGNILSC 130
+N + G+ R+ S +D RR+ GA NVF+ NLD +D K +YDTFS FGNI+S
Sbjct 90 MNMVKLYGKALRLNKSAQD---RRNFDVGA-NVFLGNLDPDVDEKTIYDTFSAFGNIISA 145
Query 131 KVGVD-EFNRSRGYGFVHYETEASARSAI 158
K+ D E SRG+GFV ++T ++ +A+
Sbjct 146 KIMRDPETGLSRGFGFVSFDTFEASDAAL 174
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 47/79 (59%), Gaps = 0/79 (0%)
Query 11 VSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERA 70
V ++++G+L P+V E +++ F+A G + S ++ RD + S G+ +V++ + ++ A
Sbjct 114 VGANVFLGNLDPDVDEKTIYDTFSAFGNIISAKIMRDPETGLSRGFGFVSFDTFEASDAA 173
Query 71 LDALNFALIRGQPCRIMWS 89
L A+N I +P + ++
Sbjct 174 LAAMNGQFICNRPIHVSYA 192
Score = 37.0 bits (84), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 32/58 (55%), Gaps = 1/58 (1%)
Query 103 VFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFNRS-RGYGFVHYETEASARSAIE 159
+++ NLD +D+ L++ F G + + V D+ + +GYGFV + E A A++
Sbjct 31 LYIGNLDSQVDDDLLWELFVQCGPVRTVSVPRDKLTGNHQGYGFVEFRNEVDADYALK 88
> ath:AT1G34140 PAB1; PAB1 (POLY(A) BINDING PROTEIN 1); RNA binding
/ translation initiation factor
Length=407
Score = 105 bits (261), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 49/87 (56%), Positives = 64/87 (73%), Gaps = 0/87 (0%)
Query 71 LDALNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSC 130
++ LN+ ++G+P RIM+S+RDPS R SG GNVFVKNLD+SIDNK L D FS FG +LSC
Sbjct 1 MEVLNYCKLKGKPMRIMFSERDPSNRMSGRGNVFVKNLDESIDNKQLCDMFSAFGKVLSC 60
Query 131 KVGVDEFNRSRGYGFVHYETEASARSA 157
KV D S+GYGFV + ++ S +A
Sbjct 61 KVARDASGVSKGYGFVQFYSDLSVYTA 87
Score = 72.4 bits (176), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 48/164 (29%), Positives = 79/164 (48%), Gaps = 18/164 (10%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+ +L T+A L +F G +TS V +D +S + +VN++ A A++
Sbjct 120 NVYVKNLVETATDADLKRLFGEFGEITSAVVMKDG-EGKSRRFGFVNFEKAEAAVTAIEK 178
Query 74 LNFALIRGQPCRIMWSQRDPS-----------------LRRSGAGNVFVKNLDKSIDNKA 116
+N ++ + + +QR + ++ N++VKNLD S+DN
Sbjct 179 MNGVVVDEKELHVGRAQRKTNRTEDLKAKFELEKIIRDMKTRKGMNLYVKNLDDSVDNTK 238
Query 117 LYDTFSLFGNILSCKVGVDEFNRSRGYGFVHYETEASARSAIEK 160
L + FS FG I SCKV V S+G GFV + T A A+ K
Sbjct 239 LEELFSEFGTITSCKVMVHSNGISKGVGFVEFSTSEEASKAMLK 282
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 48/148 (32%), Positives = 75/148 (50%), Gaps = 2/148 (1%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++++ +L + L ++F+A G V S +V RD+ S S GY +V + S A +
Sbjct 32 NVFVKNLDESIDNKQLCDMFSAFGKVLSCKVARDA-SGVSKGYGFVQFYSDLSVYTACNF 90
Query 74 LNFALIRGQPCRIM-WSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKV 132
N LIR Q + + R + NV+VKNL ++ + L F FG I S V
Sbjct 91 HNGTLIRNQHIHVCPFVSRGQWDKSRVFTNVYVKNLVETATDADLKRLFGEFGEITSAVV 150
Query 133 GVDEFNRSRGYGFVHYETEASARSAIEK 160
D +SR +GFV++E +A +AIEK
Sbjct 151 MKDGEGKSRRFGFVNFEKAEAAVTAIEK 178
> sce:YFR023W PES4; Pes4p
Length=611
Score = 103 bits (257), Expect = 2e-22, Method: Composition-based stats.
Identities = 58/148 (39%), Positives = 91/148 (61%), Gaps = 8/148 (5%)
Query 15 LYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDAL 74
L+IGDLH VTE L +F S +VC DSV+++SLG+ Y+N++ +AE+A++ L
Sbjct 93 LFIGDLHETVTEETLKGIFKKYPSFVSAKVCLDSVTKKSLGHGYLNFEDKEEAEKAMEEL 152
Query 75 NFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNL---DKSIDNKALYDTFSLFGNILSCK 131
N+ + G+ RIM S R+ + R++ NVF NL + + + YDTFS +G ILSCK
Sbjct 153 NYTKVNGKEIRIMPSLRNTTFRKNFGTNVFFSNLPLNNPLLTTRVFYDTFSRYGKILSCK 212
Query 132 VGVDEFNRSRGYGFVHYETEASARSAIE 159
+ + + GFV++E E +AR+ I+
Sbjct 213 L-----DSRKDIGFVYFEDEKTARNVIK 235
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 34/141 (24%), Positives = 65/141 (46%), Gaps = 9/141 (6%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
S++I +L T + F+ VGP+ SI + + ++ +A+V Y++ D+E+A+
Sbjct 304 SIFIKNLPTITTRDDILNFFSEVGPIKSIYL--SNATKVKYLWAFVTYKNSSDSEKAIKR 361
Query 74 LNFALIRGQPCRIMWSQ----RDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS 129
N RG+ + +Q R + +F++NL ++ NK NI
Sbjct 362 YNNFYFRGKKLLVTRAQDKEERAKFIESQKISTLFLENL-SAVCNKEFLKYLCHQENIRP 420
Query 130 CKVGVDEF--NRSRGYGFVHY 148
K+ +D + N S GF+ +
Sbjct 421 FKIQIDGYDENSSTYSGFIKF 441
> bbo:BBOV_III006220 17.m07552; RNA recognition motif domaining
containing protein; K12831 splicing factor 3B subunit 4
Length=280
Score = 102 bits (254), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 52/146 (35%), Positives = 88/146 (60%), Gaps = 2/146 (1%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
+LY+G++ +V E +L+E F VGPV + + RD V+ GYA+V + + DA+ A+
Sbjct 19 TLYVGNVDTQVDEELLWEFFVQVGPVKHLHIPRDKVTGHHQGYAFVEFDTDDDADYAIRI 78
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVG 133
LNF + +P R+ + RD GA N+F+ NLD +D+K L+DTF+ FGN++S V
Sbjct 79 LNFVKLYNKPLRLNKASRDKQTFEIGA-NLFIGNLDPDVDDKQLHDTFASFGNVISANVV 137
Query 134 VD-EFNRSRGYGFVHYETEASARSAI 158
D + + + FV Y++ ++ +A+
Sbjct 138 RDGDATDRKAFAFVSYDSFEASDAAL 163
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 49/92 (53%), Gaps = 1/92 (1%)
Query 11 VSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERA 70
+ +L+IG+L P+V + L + F + G V S V RD + +A+V+Y S ++ A
Sbjct 103 IGANLFIGNLDPDVDDKQLHDTFASFGNVISANVVRDGDATDRKAFAFVSYDSFEASDAA 162
Query 71 LDALNFALIRGQPCRIMWS-QRDPSLRRSGAG 101
L A+N I +P + ++ ++D R G+
Sbjct 163 LAAMNGQFICNKPIHVSYAYKKDTKGERHGSA 194
> sce:YHR015W MIP6; Mip6p
Length=659
Score = 102 bits (254), Expect = 6e-22, Method: Composition-based stats.
Identities = 53/150 (35%), Positives = 86/150 (57%), Gaps = 8/150 (5%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
SL+IG+L VTE ML ++F S +VCRD ++++SLGY Y+N++ DAE A
Sbjct 112 SLFIGNLKSTVTEEMLRKIFKRYQSFESAKVCRDFLTKKSLGYGYLNFKDKNDAESARKE 171
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNL---DKSIDNKALYDTFSLFGNILSC 130
N+ + GQ +IM S ++ R++ NVF NL + + ++ Y +GN+LSC
Sbjct 172 FNYTVFFGQEVKIMPSMKNTLFRKNIGTNVFFSNLPLENPQLTTRSFYLIMIEYGNVLSC 231
Query 131 KVGVDEFNRSRGYGFVHYETEASARSAIEK 160
+ R + GFV+++ + SAR+ I+K
Sbjct 232 LL-----ERRKNIGFVYFDNDISARNVIKK 256
> ath:AT2G18510 emb2444 (embryo defective 2444); RNA binding /
nucleic acid binding / nucleotide binding; K12831 splicing
factor 3B subunit 4
Length=363
Score = 102 bits (253), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 55/148 (37%), Positives = 90/148 (60%), Gaps = 3/148 (2%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+G L +++E +L+E+F GPV ++ V +D V+ Y ++ Y+S DA+ A+
Sbjct 26 TVYVGGLDAQLSEELLWELFVQAGPVVNVYVPKDRVTNLHQNYGFIEYRSEEDADYAIKV 85
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILS-CKV 132
LN + G+P R+ + +D GA N+F+ NLD +D K LYDTFS FG I S K+
Sbjct 86 LNMIKLHGKPIRVNKASQDKKSLDVGA-NLFIGNLDPDVDEKLLYDTFSAFGVIASNPKI 144
Query 133 GVD-EFNRSRGYGFVHYETEASARSAIE 159
D + SRG+GF+ Y++ ++ +AIE
Sbjct 145 MRDPDTGNSRGFGFISYDSFEASDAAIE 172
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 42/65 (64%), Gaps = 1/65 (1%)
Query 11 VSPSLYIGDLHPEVTEAMLFEVFNAVGPVTS-IRVCRDSVSRRSLGYAYVNYQSLGDAER 69
V +L+IG+L P+V E +L++ F+A G + S ++ RD + S G+ +++Y S ++
Sbjct 110 VGANLFIGNLDPDVDEKLLYDTFSAFGVIASNPKIMRDPDTGNSRGFGFISYDSFEASDA 169
Query 70 ALDAL 74
A++++
Sbjct 170 AIESM 174
> xla:734673 hypothetical protein MGC116464
Length=391
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 56/148 (37%), Positives = 85/148 (57%), Gaps = 3/148 (2%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
S+YIG+L +V E +L+E GPV S+ + RD VS GY +V +++ DA+ AL
Sbjct 12 SIYIGNLDSQVNEEILWECMLQAGPVLSVNMPRDKVSGFHQGYGFVEFKTEEDADYALKV 71
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSC-KV 132
+N + +P R + D + GA N+FV NL +D K L+DTFS FGN++S K+
Sbjct 72 MNMIKLYNKPIRCNKATTDKKIHDVGA-NLFVGNLAPEVDEKMLFDTFSQFGNLISTPKI 130
Query 133 GVD-EFNRSRGYGFVHYETEASARSAIE 159
D E S+G+ FV +++ A AI+
Sbjct 131 MKDPESGGSKGFAFVSFDSFDGADHAIQ 158
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 53/92 (57%), Gaps = 2/92 (2%)
Query 11 VSPSLYIGDLHPEVTEAMLFEVFNAVGPVTSI-RVCRDSVSRRSLGYAYVNYQSLGDAER 69
V +L++G+L PEV E MLF+ F+ G + S ++ +D S S G+A+V++ S A+
Sbjct 96 VGANLFVGNLAPEVDEKMLFDTFSQFGNLISTPKIMKDPESGGSKGFAFVSFDSFDGADH 155
Query 70 ALDALNFALIRGQPCRIMWS-QRDPSLRRSGA 100
A+ ++N + + ++ ++D + R G+
Sbjct 156 AIQSMNGQFFYNKQIVVQYAYKKDSNGERYGS 187
Score = 37.7 bits (86), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 39/67 (58%), Gaps = 1/67 (1%)
Query 94 SLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSCKVGVDEFNR-SRGYGFVHYETEA 152
S +R+ ++++ NLD ++ + L++ G +LS + D+ + +GYGFV ++TE
Sbjct 4 SEQRNQEASIYIGNLDSQVNEEILWECMLQAGPVLSVNMPRDKVSGFHQGYGFVEFKTEE 63
Query 153 SARSAIE 159
A A++
Sbjct 64 DADYALK 70
> xla:379792 sf3b4, MGC52742, spx; splicing factor 3b, subunit
4, 49kDa
Length=377
Score = 96.3 bits (238), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 54/148 (36%), Positives = 87/148 (58%), Gaps = 3/148 (2%)
Query 14 SLYIGDLHPEVTEAMLFEVFNAVGPVTSIRVCRDSVSRRSLGYAYVNYQSLGDAERALDA 73
++Y+G L +V+E +L+E+F GPV + + +D V+ + GY +V + S DA+ A+
Sbjct 14 TVYVGGLDEKVSEPLLWELFLQAGPVVNTHMPKDRVTGQHQGYGFVEFLSEEDADYAIKI 73
Query 74 LNFALIRGQPCRIMWSQRDPSLRRSGAGNVFVKNLDKSIDNKALYDTFSLFGNILSC-KV 132
+N + G+P R+ + GA N+F+ NLD ID K LYDTFS FG IL K+
Sbjct 74 MNMIKLYGKPIRVNKASAHNKNLDVGA-NIFIGNLDPEIDEKLLYDTFSAFGVILQTPKI 132
Query 133 GVD-EFNRSRGYGFVHYETEASARSAIE 159
D + S+GY F++Y + ++ +AIE
Sbjct 133 MRDPDTGNSKGYAFINYASFDASDAAIE 160
Lambda K H
0.320 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3712313100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40