bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7499_orf1
Length=181
Score E
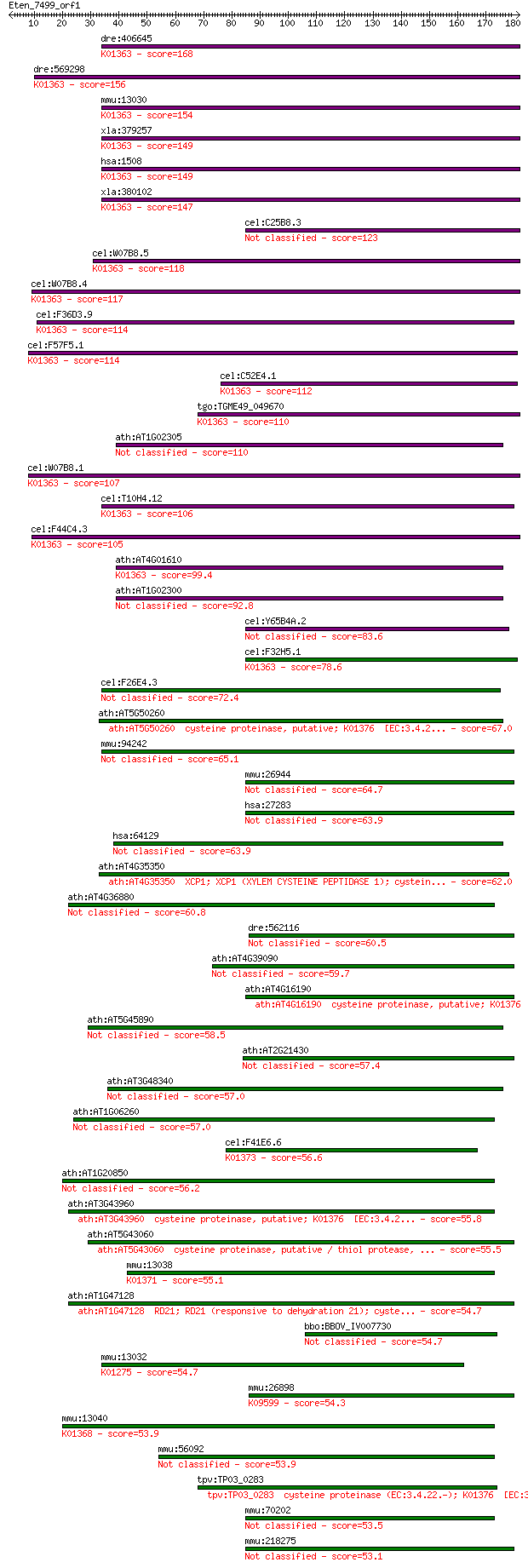
Sequences producing significant alignments: (Bits) Value
dre:406645 ctsba, MGC55862, ctsb, id:ibd1201, wu:fa13g05, wu:f... 168 7e-42
dre:569298 ctsbb; capthepsin B, b; K01363 cathepsin B [EC:3.4.... 156 4e-38
mmu:13030 Ctsb, CB; cathepsin B (EC:3.4.22.1); K01363 cathepsi... 154 1e-37
xla:379257 ctsb, MGC53360, apps, cpsb; cathepsin B (EC:3.4.22.... 149 3e-36
hsa:1508 CTSB, APPS, CPSB; cathepsin B (EC:3.4.22.1); K01363 c... 149 6e-36
xla:380102 cg10992; hypothetical protein MGC52983; K01363 cath... 147 3e-35
cel:C25B8.3 cpr-6; Cysteine PRotease related family member (cp... 123 4e-28
cel:W07B8.5 cpr-5; Cysteine PRotease related family member (cp... 118 1e-26
cel:W07B8.4 hypothetical protein; K01363 cathepsin B [EC:3.4.2... 117 2e-26
cel:F36D3.9 cpr-2; Cysteine PRotease related family member (cp... 114 2e-25
cel:F57F5.1 hypothetical protein; K01363 cathepsin B [EC:3.4.2... 114 2e-25
cel:C52E4.1 cpr-1; Cysteine PRotease related family member (cp... 112 7e-25
tgo:TGME49_049670 cysteine proteinase, putative (EC:3.4.22.1);... 110 2e-24
ath:AT1G02305 cathepsin B-like cysteine protease, putative 110 3e-24
cel:W07B8.1 hypothetical protein; K01363 cathepsin B [EC:3.4.2... 107 2e-23
cel:T10H4.12 cpr-3; Cysteine PRotease related family member (c... 106 4e-23
cel:F44C4.3 cpr-4; Cysteine PRotease related family member (cp... 105 7e-23
ath:AT4G01610 cathepsin B-like cysteine protease, putative; K0... 99.4 6e-21
ath:AT1G02300 cathepsin B-like cysteine protease, putative 92.8 5e-19
cel:Y65B4A.2 hypothetical protein 83.6 4e-16
cel:F32H5.1 hypothetical protein; K01363 cathepsin B [EC:3.4.2... 78.6 1e-14
cel:F26E4.3 hypothetical protein 72.4 9e-13
ath:AT5G50260 cysteine proteinase, putative; K01376 [EC:3.4.2... 67.0 3e-11
mmu:94242 Tinagl1, 1110021J17Rik, AZ-1, AZ1, Arg1, Lcn7, TARP,... 65.1 1e-10
mmu:26944 Tinag, AI452335, TIN-ag; tubulointerstitial nephriti... 64.7 1e-10
hsa:27283 TINAG, TIN-AG; tubulointerstitial nephritis antigen 63.9 2e-10
hsa:64129 TINAGL1, ARG1, LCN7, LIECG3, TINAGRP; tubulointersti... 63.9 3e-10
ath:AT4G35350 XCP1; XCP1 (XYLEM CYSTEINE PEPTIDASE 1); cystein... 62.0 1e-09
ath:AT4G36880 CP1; CP1 (CYSTEINE PROTEINASE1); cysteine-type e... 60.8 2e-09
dre:562116 tinagl1, si:dkey-158b13.1; tubulointerstitial nephr... 60.5 3e-09
ath:AT4G39090 RD19; RD19 (RESPONSIVE TO DEHYDRATION 19); cyste... 59.7 6e-09
ath:AT4G16190 cysteine proteinase, putative; K01376 [EC:3.4.2... 58.5 1e-08
ath:AT5G45890 SAG12; SAG12 (SENESCENCE-ASSOCIATED GENE 12); cy... 58.5 1e-08
ath:AT2G21430 cysteine proteinase A494, putative / thiol prote... 57.4 3e-08
ath:AT3G48340 cysteine-type endopeptidase/ cysteine-type pepti... 57.0 3e-08
ath:AT1G06260 cysteine proteinase, putative 57.0 3e-08
cel:F41E6.6 tag-196; Temporarily Assigned Gene name family mem... 56.6 5e-08
ath:AT1G20850 XCP2; XCP2 (xylem cysteine peptidase 2); cystein... 56.2 6e-08
ath:AT3G43960 cysteine proteinase, putative; K01376 [EC:3.4.2... 55.8 8e-08
ath:AT5G43060 cysteine proteinase, putative / thiol protease, ... 55.5 1e-07
mmu:13038 Ctsk, AI323530, MMS10-Q, Ms10q, catK; cathepsin K (E... 55.1 1e-07
ath:AT1G47128 RD21; RD21 (responsive to dehydration 21); cyste... 54.7 2e-07
bbo:BBOV_IV007730 23.m06535; cysteine protease 2 54.7
mmu:13032 Ctsc, AI047818, DPP1, DPPI; cathepsin C (EC:3.4.14.1... 54.7 2e-07
mmu:26898 Ctsj, CATP, CatRLP, Ctsp, MGC117662; cathepsin J; K0... 54.3 3e-07
mmu:13040 Ctss; cathepsin S (EC:3.4.22.27); K01368 cathepsin S... 53.9 3e-07
mmu:56092 Cts7, AA408230, AA408952, CTS1, Epcs24, Epcs71; cath... 53.9 3e-07
tpv:TP03_0283 cysteine proteinase (EC:3.4.22.-); K01376 [EC:3... 53.5 4e-07
mmu:70202 Ctsll3, 2310051M13Rik; cathepsin L-like 3 (EC:3.4.22... 53.5 4e-07
mmu:218275 MGC60730; cDNA sequence BC051665 53.1 4e-07
> dre:406645 ctsba, MGC55862, ctsb, id:ibd1201, wu:fa13g05, wu:fb34e12,
zgc:55862, zgc:65809, zgc:77181; cathepsin B, a; K01363
cathepsin B [EC:3.4.22.1]
Length=330
Score = 168 bits (426), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 84/149 (56%), Positives = 106/149 (71%), Gaps = 6/149 (4%)
Query 34 IHTINNMKTTWEAGENFGPHITSDYIRNLCGA-LKTPLSKKLPIKDLSKEVHDLPIEFDA 92
++ IN TTW AG NF + Y++ LCG LK P KLP+ E LP FDA
Sbjct 30 VNFINKANTTWTAGHNF-RDVDYSYVKKLCGTFLKGP---KLPVMVQYTEGLKLPKNFDA 85
Query 93 RKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCDSC 152
R++W + CP+L E+RDQG CGSCWAFGAAEA++DR+CI + K V IS++DLLTCCDSC
Sbjct 86 REQWPN-CPTLKEIRDQGSCGSCWAFGAAEAISDRVCIHSDAKVSVEISSQDLLTCCDSC 144
Query 153 GFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
G GCNGGYP +AW+F+ T+G+VTGG YNS
Sbjct 145 GMGCNGGYPSAAWDFWATEGLVTGGLYNS 173
> dre:569298 ctsbb; capthepsin B, b; K01363 cathepsin B [EC:3.4.22.1]
Length=326
Score = 156 bits (394), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 79/173 (45%), Positives = 112/173 (64%), Gaps = 7/173 (4%)
Query 10 KLIIFGVLIAMVFTMPKNSMFQSHIHTINNMKTTWEAGENFGPHITSDYIRNLCG-ALKT 68
++ +F VL+++ P+ I IN ++TW AG NF ++ +Y+++LCG LK
Sbjct 3 RVCVF-VLLSVTCARPQLHTHDEMISFINAARSTWTAGVNFD-NVPKEYLKSLCGTVLKG 60
Query 69 PLSKKLPIKDLSKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRI 128
P +LP LP FD R +W + C +L ++RDQG CGSCWAFGA E+++DRI
Sbjct 61 P---RLPHTVKHSTNVKLPDSFDLRDQWPN-CKTLSQIRDQGSCGSCWAFGAVESISDRI 116
Query 129 CIATKGKNQVRISTEDLLTCCDSCGFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
CI +KGK IS EDLL+CCD CGFGC+GG+P AW++++ G+VTGG YNS
Sbjct 117 CIHSKGKQSPEISAEDLLSCCDQCGFGCSGGFPAEAWDYWRRSGLVTGGLYNS 169
> mmu:13030 Ctsb, CB; cathepsin B (EC:3.4.22.1); K01363 cathepsin
B [EC:3.4.22.1]
Length=339
Score = 154 bits (390), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 79/149 (53%), Positives = 100/149 (67%), Gaps = 5/149 (3%)
Query 34 IHTINNMKTTWEAGENFGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEVHDLPIEFDAR 93
I+ IN TTW+AG NF ++ Y++ LCG + KLP + E DLP FDAR
Sbjct 31 INYINKQNTTWQAGRNF-YNVDISYLKKLCGTVLG--GPKLPGRVAFGEDIDLPETFDAR 87
Query 94 KEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCD-SC 152
++W S CP++ ++RDQG CGSCWAFGA EA++DR CI T G+ V +S EDLLTCC C
Sbjct 88 EQW-SNCPTIGQIRDQGSCGSCWAFGAVEAISDRTCIHTNGRVNVEVSAEDLLTCCGIQC 146
Query 153 GFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
G GCNGGYP AW F+ KG+V+GG YNS
Sbjct 147 GDGCNGGYPSGAWSFWTKKGLVSGGVYNS 175
> xla:379257 ctsb, MGC53360, apps, cpsb; cathepsin B (EC:3.4.22.1);
K01363 cathepsin B [EC:3.4.22.1]
Length=333
Score = 149 bits (377), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 76/151 (50%), Positives = 101/151 (66%), Gaps = 9/151 (5%)
Query 34 IHTINNMKTTWEAGENFGPHITSDYIRNLCGAL-KTP-LSKKLPIKDLSKEVHDLPIEFD 91
++ IN + TTW+AG NF + Y++ LCG L K P L K+ D +LP FD
Sbjct 31 VNYINKVNTTWKAGHNFA-NADLHYVKRLCGTLLKGPQLQKRFGFADGL----ELPDSFD 85
Query 92 ARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCC-D 150
+R W + CP++ E+RDQG CGSCWAFGA EA++DR+C+ T GK V +S EDLL+CC D
Sbjct 86 SRAAWPN-CPTIREIRDQGSCGSCWAFGAVEAISDRVCVHTNGKVNVEVSAEDLLSCCGD 144
Query 151 SCGFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
CG GCNGGYP AW+F+ G+V+GG Y+S
Sbjct 145 ECGMGCNGGYPSGAWQFWTETGLVSGGLYDS 175
> hsa:1508 CTSB, APPS, CPSB; cathepsin B (EC:3.4.22.1); K01363
cathepsin B [EC:3.4.22.1]
Length=339
Score = 149 bits (375), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 75/149 (50%), Positives = 96/149 (64%), Gaps = 5/149 (3%)
Query 34 IHTINNMKTTWEAGENFGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEVHDLPIEFDAR 93
++ +N TTW+AG NF ++ Y++ LCG K P + + E LP FDAR
Sbjct 31 VNYVNKRNTTWQAGHNF-YNVDMSYLKRLCGTFLG--GPKPPQRVMFTEDLKLPASFDAR 87
Query 94 KEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCDS-C 152
++W CP++ E+RDQG CGSCWAFGA EA++DRICI T V +S EDLLTCC S C
Sbjct 88 EQWPQ-CPTIKEIRDQGSCGSCWAFGAVEAISDRICIHTNAHVSVEVSAEDLLTCCGSMC 146
Query 153 GFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
G GCNGGYP AW F+ KG+V+GG Y S
Sbjct 147 GDGCNGGYPAEAWNFWTRKGLVSGGLYES 175
> xla:380102 cg10992; hypothetical protein MGC52983; K01363 cathepsin
B [EC:3.4.22.1]
Length=333
Score = 147 bits (370), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 75/151 (49%), Positives = 98/151 (64%), Gaps = 9/151 (5%)
Query 34 IHTINNMKTTWEAGENFGPHITSDYIRNLCGA-LKTP-LSKKLPIKDLSKEVHDLPIEFD 91
++ IN + TTW+AG NF + Y++ LCG L P L K+ D DLP FD
Sbjct 31 VNYINKVNTTWKAGHNFA-NADVHYVKRLCGTHLNGPQLQKRFGFADDL----DLPDSFD 85
Query 92 ARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCD- 150
+R W + CP++ E+RDQG CGSCWAFGA EA++DR+C+ T GK V +S EDLL+CC
Sbjct 86 SRAAWPN-CPTIREIRDQGSCGSCWAFGAVEAISDRVCVHTNGKVNVEVSAEDLLSCCGF 144
Query 151 SCGFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
CG GCNGGYP AW F+ G+V+GG Y+S
Sbjct 145 KCGMGCNGGYPSGAWRFWTETGLVSGGLYDS 175
> cel:C25B8.3 cpr-6; Cysteine PRotease related family member (cpr-6)
Length=379
Score = 123 bits (308), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 56/97 (57%), Positives = 70/97 (72%), Gaps = 1/97 (1%)
Query 85 DLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTED 144
D+P FD+R W C S+ +RDQ CGSCWAFGA EAM+DRICIA+ G+ QV +S +D
Sbjct 104 DIPESFDSRDNWPK-CDSIKVIRDQSSCGSCWAFGAVEAMSDRICIASHGELQVTLSADD 162
Query 145 LLTCCDSCGFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
LL+CC SCGFGCNGG P +AW ++ GIVTG Y +
Sbjct 163 LLSCCKSCGFGCNGGDPLAAWRYWVKDGIVTGSNYTA 199
> cel:W07B8.5 cpr-5; Cysteine PRotease related family member (cpr-5);
K01363 cathepsin B [EC:3.4.22.1]
Length=344
Score = 118 bits (296), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 69/165 (41%), Positives = 94/165 (56%), Gaps = 29/165 (17%)
Query 31 QSHIHTINNMKTTWEAGENFGPHITSDYIRNLCGALKTPLSKKL-------PIKD---LS 80
Q+ I +N+ + W AG P K ++KKL P KD ++
Sbjct 30 QALIDYVNSAQKLWTAGHQVIP--------------KEKITKKLMDVKYLVPHKDEDIVA 75
Query 81 KEVHD-LPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVR 139
EV D +P FDAR +W + C S+ +RDQ +CGSCWAF AAEA++DR CIA+ G
Sbjct 76 TEVSDAIPDHFDARDQWPN-CMSINNIRDQSDCGSCWAFAAAEAISDRTCIASNGAVNTL 134
Query 140 ISTEDLLTCCD---SCGFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
+S+EDLL+CC SCG GC GGYP AW+++ G+VTGG Y +
Sbjct 135 LSSEDLLSCCTGMFSCGNGCEGGYPIQAWKWWVKHGLVTGGSYET 179
> cel:W07B8.4 hypothetical protein; K01363 cathepsin B [EC:3.4.22.1]
Length=335
Score = 117 bits (293), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 67/179 (37%), Positives = 97/179 (54%), Gaps = 16/179 (8%)
Query 9 MKLI--IFGVLIAMVFTMPKNSMFQSHIHTINNMKTTWEAGENFGPHITSDY-IRNLCGA 65
+KL+ + +L A +P+N +F +HI N+ + W A H T+ + ++NL
Sbjct 2 LKLLPSLLFILAASAVVLPRNKLFINHI---NSAQKLWTA-----EHYTTPFEVKNLMKV 53
Query 66 LKTPLSKKLPIKDLSKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMT 125
IK L++ +P +D R W C S+ +RDQ CGSCWA AAEA++
Sbjct 54 EHVAAHLDKDIK-LAETADSIPDSYDVRDHWPQ-CISVNNIRDQSHCGSCWAVAAAEAIS 111
Query 126 DRICIATKGKNQVRISTEDLLTCCD---SCGFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
DR CIA+ G +S ED+LTCC +CG GC GGYP AW ++ G+VTGG + S
Sbjct 112 DRTCIASNGDVNTLLSAEDILTCCTGKFNCGDGCEGGYPIQAWRYWVKNGLVTGGSFES 170
> cel:F36D3.9 cpr-2; Cysteine PRotease related family member (cpr-2);
K01363 cathepsin B [EC:3.4.22.1]
Length=344
Score = 114 bits (285), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 60/170 (35%), Positives = 91/170 (53%), Gaps = 3/170 (1%)
Query 11 LIIFGVLIAMVFTMPKNSMFQSHIHTINNMKTTWEAGENFGPHITSDYIRNLCGALKTPL 70
L + A + +P+ Q + IN+ +T++ EN+ + R++ P
Sbjct 27 LFVITFAFAPIRPIPEELTGQDLVDHINSAASTFQT-ENYAVTHEKMHTRSMHEKFNAPF 85
Query 71 SKKLPIKDLSKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICI 130
+ + + P+ FDAR W C S+ +R+Q CGSCWAF AE ++DR CI
Sbjct 86 PDEFRATEREFVLDATPLNFDARTRWPQ-CKSMKLIREQSNCGSCWAFSTAEVISDRTCI 144
Query 131 ATKGKNQVRISTEDLLTCCD-SCGFGCNGGYPQSAWEFFKTKGIVTGGPY 179
A+ G Q IS DLLTCC SCG GC+GG+P A++++ +G+VTGG Y
Sbjct 145 ASNGTQQPIISPTDLLTCCGMSCGEGCDGGFPYRAFQWWARRGVVTGGDY 194
> cel:F57F5.1 hypothetical protein; K01363 cathepsin B [EC:3.4.22.1]
Length=400
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 71/195 (36%), Positives = 104/195 (53%), Gaps = 25/195 (12%)
Query 8 KMKLIIFGVLIAMVFTMPKNSMF---------------QSHIHTINNMKTTWEAGENFGP 52
KMK I +L+ +V N Q + +N ++T+++A G
Sbjct 49 KMKTAIAALLVGLVAVNAYNVEVKHGDAIPVEAQMLRGQELVDYVNKVQTSFKA--ELGS 106
Query 53 HITS--DYIR-NLCGALKTPLSKKLPIKDLSK-EVHD--LPIEFDARKEWGSICPSLLEV 106
+ +S D I+ L GA + ++ + +++ EV D +P FD+R W + CPS+ ++
Sbjct 107 YFSSYPDTIKKQLMGAKMVEIPEEYRVFEMTHPEVEDAAVPDSFDSRTAWPN-CPSISKI 165
Query 107 RDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCD-SCGFGCNGGYPQSAW 165
RDQ CGSCWA AAE ++DRICIA+ K + IS +D+ CC CG GCNGGYP AW
Sbjct 166 RDQSSCGSCWAVSAAETISDRICIASNAKTILSISADDINACCGMVCGNGCNGGYPIEAW 225
Query 166 EFFKTKGIVTGGPYN 180
+ KG VTGG Y
Sbjct 226 RHYVKKGYVTGGSYQ 240
> cel:C52E4.1 cpr-1; Cysteine PRotease related family member (cpr-1);
K01363 cathepsin B [EC:3.4.22.1]
Length=329
Score = 112 bits (280), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 57/108 (52%), Positives = 73/108 (67%), Gaps = 4/108 (3%)
Query 76 IKDLSKEV--HDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATK 133
I+ +EV +P FD+R +W S C S+ +RDQ CGSCWAFGAAE ++DR CI TK
Sbjct 73 IRATEQEVVLASVPATFDSRTQW-SECKSIKLIRDQATCGSCWAFGAAEMISDRTCIETK 131
Query 134 GKNQVRISTEDLLTCC-DSCGFGCNGGYPQSAWEFFKTKGIVTGGPYN 180
G Q IS +DLL+CC SCG GC GGYP A ++ +KG+VTGG Y+
Sbjct 132 GAQQPIISPDDLLSCCGSSCGNGCEGGYPIQALRWWDSKGVVTGGDYH 179
> tgo:TGME49_049670 cysteine proteinase, putative (EC:3.4.22.1);
K01363 cathepsin B [EC:3.4.22.1]
Length=572
Score = 110 bits (276), Expect = 2e-24, Method: Composition-based stats.
Identities = 49/117 (41%), Positives = 71/117 (60%), Gaps = 3/117 (2%)
Query 68 TPLSKKLPIKDLSKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDR 127
TP LP K+ +P FDAR + + + VRDQG+CGSCWAF + EA DR
Sbjct 259 TPKGMPLPAKEFENATEPVPAHFDARTAFPACKDVVGHVRDQGDCGSCWAFASTEAFNDR 318
Query 128 ICIATKGKNQVRISTEDLLTCCDS---CGFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
+CI ++GK + +S + +CC++ FGCNGG P AW +F+ KG+VTGG +++
Sbjct 319 LCIRSQGKGLMPLSAQHTTSCCNAIHCASFGCNGGQPGMAWRWFERKGVVTGGDFDA 375
> ath:AT1G02305 cathepsin B-like cysteine protease, putative
Length=362
Score = 110 bits (275), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 64/141 (45%), Positives = 83/141 (58%), Gaps = 8/141 (5%)
Query 39 NMKTTWEAGEN--FGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEVH-DLPIEFDARKE 95
N W+A N F +++ R L G TP ++ L + +S ++ LP EFDAR
Sbjct 57 NPNAGWKASFNDRFANATVAEFKR-LLGVKPTPKTEFLGVPIVSHDISLKLPKEFDARTA 115
Query 96 WGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCDS-CGF 154
W S C S+ + DQG CGSCWAFGA E+++DR CI K V +S DLL CC CG
Sbjct 116 W-SQCTSIGRILDQGHCGSCWAFGAVESLSDRFCI--KYNMNVSLSVNDLLACCGFLCGQ 172
Query 155 GCNGGYPQSAWEFFKTKGIVT 175
GCNGGYP +AW +FK G+VT
Sbjct 173 GCNGGYPIAAWRYFKHHGVVT 193
> cel:W07B8.1 hypothetical protein; K01363 cathepsin B [EC:3.4.22.1]
Length=335
Score = 107 bits (268), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 65/177 (36%), Positives = 93/177 (52%), Gaps = 9/177 (5%)
Query 8 KMKLIIFGVLIAMVFTMPKNSMFQSHIHTINNMKTTWEAGENFGPHITSDYIRNLCGALK 67
K+ + + GVL P S IH +N+ KTTW AG P ++ + +
Sbjct 3 KILICLIGVLFQADGVPP--SEIDRIIHYVNSQKTTWTAGI---PALSRNSMLKTLVTDA 57
Query 68 TPLSKKLPIKDLSKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDR 127
+ K+ +S+ DL FDAR+ W C S+ ++ D EC + WAF AAE+M+DR
Sbjct 58 ATIGFKIQNFGVSQANSDLSPSFDARERWPE-CMSIPQINDISECKTSWAFAAAESMSDR 116
Query 128 ICIATKGKNQVRISTEDLLTCCD---SCGFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
+CI + G +S E+LL+CC SCG GC GG P AW++ + GI TGG Y S
Sbjct 117 LCINSGGFKNTILSAEELLSCCTGMFSCGEGCEGGNPFKAWQYIQKHGIPTGGSYES 173
> cel:T10H4.12 cpr-3; Cysteine PRotease related family member
(cpr-3); K01363 cathepsin B [EC:3.4.22.1]
Length=370
Score = 106 bits (265), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 59/155 (38%), Positives = 86/155 (55%), Gaps = 14/155 (9%)
Query 34 IHTINNMKTTWEAGENFGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEV--------HD 85
+ +N ++T+W A N + + + PL K D++ E+
Sbjct 36 VDHVNTVQTSWVAEHNEISEFEMKF-KVMDVKFAEPLEKD---SDVASELFVRGEIVPEP 91
Query 86 LPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDL 145
LP FDAR++W C ++ +R+Q CGSCWAFGAAE ++DR+CI + G Q IS ED+
Sbjct 92 LPDTFDAREKWPD-CNTIKLIRNQATCGSCWAFGAAEVISDRVCIQSNGTQQPVISVEDI 150
Query 146 LTCC-DSCGFGCNGGYPQSAWEFFKTKGIVTGGPY 179
L+CC +CG+GC GGY A F+ + G VTGG Y
Sbjct 151 LSCCGTTCGYGCKGGYSIEALRFWASSGAVTGGDY 185
> cel:F44C4.3 cpr-4; Cysteine PRotease related family member (cpr-4);
K01363 cathepsin B [EC:3.4.22.1]
Length=335
Score = 105 bits (263), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 63/176 (35%), Positives = 93/176 (52%), Gaps = 4/176 (2%)
Query 9 MKLIIFGVLIAMV--FTMPKNSMFQSHI-HTINNMKTTWEAGENFGPHITSDYIRNLCGA 65
MK +I L+A+ +P Q I +N+ ++ W+A I R +
Sbjct 1 MKYLILAALVAVTAGLVIPLVPKTQEAITEYVNSKQSLWKAEIPKDITIEQVKKRLMRTE 60
Query 66 LKTPLSKKLPIKDLSKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMT 125
P + + + +P FDAR +W + C S+ +RDQ +CGSCWAF AAEA +
Sbjct 61 FVAPHTPDVEVVKHDINEDTIPATFDARTQWPN-CMSINNIRDQSDCGSCWAFAAAEAAS 119
Query 126 DRICIATKGKNQVRISTEDLLTCCDSCGFGCNGGYPQSAWEFFKTKGIVTGGPYNS 181
DR CIA+ G +S ED+L+CC +CG+GC GGYP +AW++ G TGG Y +
Sbjct 120 DRFCIASNGAVNTLLSAEDVLSCCSNCGYGCEGGYPINAWKYLVKSGFCTGGSYEA 175
> ath:AT4G01610 cathepsin B-like cysteine protease, putative;
K01363 cathepsin B [EC:3.4.22.1]
Length=359
Score = 99.4 bits (246), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 60/141 (42%), Positives = 79/141 (56%), Gaps = 8/141 (5%)
Query 39 NMKTTWEAGEN--FGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEVH-DLPIEFDARKE 95
N W+A N F +++ R L G TP L + +S + LP FDAR
Sbjct 54 NPNAGWKAAINDRFSNATVAEFKR-LLGVKPTPKKHFLGVPIVSHDPSLKLPKAFDARTA 112
Query 96 WGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCD-SCGF 154
W C S+ + DQG CGSCWAFGA E+++DR CI G N + +S DLL CC CG
Sbjct 113 WPQ-CTSIGNILDQGHCGSCWAFGAVESLSDRFCIQF-GMN-ISLSVNDLLACCGFRCGD 169
Query 155 GCNGGYPQSAWEFFKTKGIVT 175
GC+GGYP +AW++F G+VT
Sbjct 170 GCDGGYPIAAWQYFSYSGVVT 190
> ath:AT1G02300 cathepsin B-like cysteine protease, putative
Length=379
Score = 92.8 bits (229), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 59/161 (36%), Positives = 81/161 (50%), Gaps = 28/161 (17%)
Query 39 NMKTTWEAGEN--FGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEVH-DLPIEFDARKE 95
N W+A N F +++ R L G ++TP + L + + ++ LP EFDAR
Sbjct 54 NPNAGWKAAFNDRFANATVAEFKR-LLGVIQTPKTAYLGVPIVRHDLSLKLPKEFDARTA 112
Query 96 WGSICPSLLEVRD--------------------QGECGSCWAFGAAEAMTDRICIATKGK 135
W S C S+ + G CGSCWAFGA E+++DR CI K
Sbjct 113 W-SHCTSIRRILVGYILNNVLLWSTITLWFWFLLGHCGSCWAFGAVESLSDRFCI--KYN 169
Query 136 NQVRISTEDLLTCCDS-CGFGCNGGYPQSAWEFFKTKGIVT 175
V +S D++ CC CGFGCNGG+P AW +FK G+VT
Sbjct 170 LNVSLSANDVIACCGLLCGFGCNGGFPMGAWLYFKYHGVVT 210
> cel:Y65B4A.2 hypothetical protein
Length=421
Score = 83.6 bits (205), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 42/93 (45%), Positives = 59/93 (63%), Gaps = 2/93 (2%)
Query 85 DLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTED 144
D+P FDAR++W + CPS+ V +QG CGSC+A AA +DR CI + G + +S ED
Sbjct 137 DVPKNFDARQKWPN-CPSISNVPNQGGCGSCFAVAAAGVASDRACIHSNGTFKSLLSEED 195
Query 145 LLTCCDSCGFGCNGGYPQSAWEFFKTKGIVTGG 177
++ CC CG C GG P A ++ +G+VTGG
Sbjct 196 IIGCCSVCG-NCYGGDPLKALTYWVNQGLVTGG 227
> cel:F32H5.1 hypothetical protein; K01363 cathepsin B [EC:3.4.22.1]
Length=356
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 40/102 (39%), Positives = 60/102 (58%), Gaps = 7/102 (6%)
Query 85 DLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTED 144
D+P FD+R++W S C + VRDQ +CGS A E +DR CIA+ G +S +D
Sbjct 91 DIPSSFDSRQKWPS-CSQIGAVRDQSDCGSAAHLVAVEIASDRTCIASNGTFNWPLSAQD 149
Query 145 LLTCCDSC------GFGCNGGYPQSAWEFFKTKGIVTGGPYN 180
L+CC G+GC+G +P+ ++++T G+ TGG YN
Sbjct 150 PLSCCVGLMSICGDGWGCDGSWPKDILKWWQTHGLCTGGNYN 191
> cel:F26E4.3 hypothetical protein
Length=452
Score = 72.4 bits (176), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 43/147 (29%), Positives = 73/147 (49%), Gaps = 14/147 (9%)
Query 34 IHTINNMKTTWEAGENFGPHITSDYIRNLCGALKTPLSKKLPIKDLSK------EVHDLP 87
+ I+ + +W A N+ ++ + R+L +K L P + + + +LP
Sbjct 131 LEKIHTGRYSWSA-RNY----SAFWGRSLSDGIKYRLGTLFPERSVQNMNEILIKPRELP 185
Query 88 IEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLT 147
FDAR +WG P + V DQG+CGS W+ +DR+ I ++G+ +S++ LL+
Sbjct 186 EHFDARDKWG---PLIHPVADQGDCGSSWSVSTTAISSDRLAIISEGRINSTLSSQQLLS 242
Query 148 CCDSCGFGCNGGYPQSAWEFFKTKGIV 174
C GC GGY AW + + G+V
Sbjct 243 CNQHRQKGCEGGYLDRAWWYIRKLGVV 269
> ath:AT5G50260 cysteine proteinase, putative; K01376 [EC:3.4.22.-]
Length=361
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 50/155 (32%), Positives = 75/155 (48%), Gaps = 25/155 (16%)
Query 33 HIHTINNMKTTWEAGEN-FGPHITSDYIRNLCGA-LKTPL----SKKLPIKDLSKEVHDL 86
HIH N +++ N FG + ++ R G+ +K KK + V+ L
Sbjct 67 HIHETNKKDKSYKLKLNKFGDMTSEEFRRTYAGSNIKHHRMFQGEKKATKSFMYANVNTL 126
Query 87 PIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVR------I 140
P D RK G++ P V++QG+CGSCWAF +A +G NQ+R +
Sbjct 127 PTSVDWRKN-GAVTP----VKNQGQCGSCWAFST--------VVAVEGINQIRTKKLTSL 173
Query 141 STEDLLTCCDSCGFGCNGGYPQSAWEFFKTKGIVT 175
S ++L+ C + GCNGG A+EF K KG +T
Sbjct 174 SEQELVDCDTNQNQGCNGGLMDLAFEFIKEKGGLT 208
> mmu:94242 Tinagl1, 1110021J17Rik, AZ-1, AZ1, Arg1, Lcn7, TARP,
Tinagl; tubulointerstitial nephritis antigen-like 1
Length=466
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 46/152 (30%), Positives = 72/152 (47%), Gaps = 11/152 (7%)
Query 34 IHTINNMKTTWEAGEN--FGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEVHD---LPI 88
I IN W+AG + F + IR G ++ P S + + ++ + LP
Sbjct 146 IKAINRGNYGWQAGNHSAFWGMTLDEGIRYRLGTIR-PSSTVMNMNEIYTVLGQGEVLPT 204
Query 89 EFDARKEWGSICPSLL-EVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLT 147
F+A ++W P+L+ E DQG C WAF A +DR+ I + G +S ++LL+
Sbjct 205 AFEASEKW----PNLIHEPLDQGNCAGSWAFSTAAVASDRVSIHSLGHMTPILSPQNLLS 260
Query 148 CCDSCGFGCNGGYPQSAWEFFKTKGIVTGGPY 179
C GC GG AW F + +G+V+ Y
Sbjct 261 CDTHHQQGCRGGRLDGAWWFLRRRGVVSDNCY 292
> mmu:26944 Tinag, AI452335, TIN-ag; tubulointerstitial nephritis
antigen
Length=475
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 34/95 (35%), Positives = 47/95 (49%), Gaps = 3/95 (3%)
Query 85 DLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTED 144
DLP F A +W L DQ C + WAF A DRI I +KG+ +S ++
Sbjct 215 DLPEIFIASYKWPGWTHGPL---DQKNCAASWAFSTASVAADRIAIQSKGRYTANLSPQN 271
Query 145 LLTCCDSCGFGCNGGYPQSAWEFFKTKGIVTGGPY 179
L++CC GCN G AW F + +G+V+ Y
Sbjct 272 LISCCAKNRHGCNSGSIDRAWWFLRKRGLVSHACY 306
> hsa:27283 TINAG, TIN-AG; tubulointerstitial nephritis antigen
Length=476
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 47/95 (49%), Gaps = 3/95 (3%)
Query 85 DLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTED 144
DLP F A +W L DQ C + WAF A DRI I +KG+ +S ++
Sbjct 216 DLPEFFVASYKWPGWTHGPL---DQKNCAASWAFSTASVAADRIAIQSKGRYTANLSPQN 272
Query 145 LLTCCDSCGFGCNGGYPQSAWEFFKTKGIVTGGPY 179
L++CC GCN G AW + + +G+V+ Y
Sbjct 273 LISCCAKNRHGCNSGSIDRAWWYLRKRGLVSHACY 307
> hsa:64129 TINAGL1, ARG1, LCN7, LIECG3, TINAGRP; tubulointerstitial
nephritis antigen-like 1
Length=436
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 43/144 (29%), Positives = 71/144 (49%), Gaps = 11/144 (7%)
Query 38 NNMKTTWEAGEN--FGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEVHD---LPIEFDA 92
+N W+AG + F + IR G ++ P S + + ++ ++ LP F+A
Sbjct 120 DNCNRCWQAGNHSAFWGMTLDEGIRYRLGTIR-PSSSVMNMHEIYTVLNPGEVLPTAFEA 178
Query 93 RKEWGSICPSLL-EVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCDS 151
++W P+L+ E DQG C WAF A +DR+ I + G +S ++LL+C
Sbjct 179 SEKW----PNLIHEPLDQGNCAGSWAFSTAAVASDRVSIHSLGHMTPVLSPQNLLSCDTH 234
Query 152 CGFGCNGGYPQSAWEFFKTKGIVT 175
GC GG AW F + +G+V+
Sbjct 235 QQQGCRGGRLDGAWWFLRRRGVVS 258
> ath:AT4G35350 XCP1; XCP1 (XYLEM CYSTEINE PEPTIDASE 1); cysteine-type
endopeptidase/ cysteine-type peptidase; K01376 [EC:3.4.22.-]
Length=355
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 47/148 (31%), Positives = 71/148 (47%), Gaps = 14/148 (9%)
Query 33 HIHTINNMKTTWEAGENFGPHITSDYIRNLCGALKTPL--SKKLPIKDLS-KEVHDLPIE 89
HI NN ++ G N +T + + L P K+ P + +++ DLP
Sbjct 81 HIDQRNNEINSYWLGLNEFADLTHEEFKGRYLGLAKPQFSRKRQPSANFRYRDITDLPKS 140
Query 90 FDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCC 149
D RK+ G++ P V+DQG+CGSCWAF A+ I T N +S ++L+ C
Sbjct 141 VDWRKK-GAVAP----VKDQGQCGSCWAFSTVAAVEGINQITT--GNLSSLSEQELIDCD 193
Query 150 DSCGFGCNGGYPQSAWEFFKTKGIVTGG 177
+ GCNGG A+++ I TGG
Sbjct 194 TTFNSGCNGGLMDYAFQYI----ISTGG 217
> ath:AT4G36880 CP1; CP1 (CYSTEINE PROTEINASE1); cysteine-type
endopeptidase/ cysteine-type peptidase
Length=376
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 45/158 (28%), Positives = 76/158 (48%), Gaps = 14/158 (8%)
Query 22 FTMPKNSMFQSHIHTINNMKTTWEAGENFGPHITSDYIRNL-CGALKTPLSKKLPIKDLS 80
F + K+++ +H NN T++ G +T+D R L GA P + K+++
Sbjct 74 FNIFKDNLRFIDLHNENNKNATYKLGLTKFTDLTNDEYRKLYLGARTEPARRIAKAKNVN 133
Query 81 KEV------HDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKG 134
++ ++P D R++ G++ P ++DQG CGSCWAF A+ I T
Sbjct 134 QKYSAAVNGKEVPETVDWRQK-GAVNP----IKDQGTCGSCWAFSTTAAVEGINKIVT-- 186
Query 135 KNQVRISTEDLLTCCDSCGFGCNGGYPQSAWEFFKTKG 172
+ +S ++L+ C S GCNGG A++F G
Sbjct 187 GELISLSEQELVDCDKSYNQGCNGGLMDYAFQFIMKNG 224
> dre:562116 tinagl1, si:dkey-158b13.1; tubulointerstitial nephritis
antigen-like 1
Length=471
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 48/94 (51%), Gaps = 3/94 (3%)
Query 86 LPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDL 145
LP F+A +W + E DQG C + WAF A +DRI I + G ++S ++L
Sbjct 200 LPSYFNAVDKWPG---KIHEPLDQGNCNASWAFSTAAVASDRISIQSMGHMTPQLSPQNL 256
Query 146 LTCCDSCGFGCNGGYPQSAWEFFKTKGIVTGGPY 179
++C GC GG AW F + +G+VT Y
Sbjct 257 ISCDTRHQDGCAGGRIDGAWWFMRRRGVVTQDCY 290
> ath:AT4G39090 RD19; RD19 (RESPONSIVE TO DEHYDRATION 19); cysteine-type
endopeptidase/ cysteine-type peptidase
Length=368
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 47/120 (39%), Positives = 65/120 (54%), Gaps = 21/120 (17%)
Query 73 KLPIKDLSKE----VHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRI 128
KLP KD +K +LP +FD R + G++ P V++QG CGSCW+F A A+
Sbjct 119 KLP-KDANKAPILPTENLPEDFDWR-DHGAVTP----VKNQGSCGSCWSFSATGALEGAN 172
Query 129 CIATKGKNQVRISTEDLLTC--------CDSCGFGCNGGYPQSAWEF-FKTKGIVTGGPY 179
+AT GK V +S + L+ C DSC GCNGG SA+E+ KT G++ Y
Sbjct 173 FLAT-GK-LVSLSEQQLVDCDHECDPEEADSCDSGCNGGLMNSAFEYTLKTGGLMKEEDY 230
> ath:AT4G16190 cysteine proteinase, putative; K01376 [EC:3.4.22.-]
Length=373
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 57/104 (54%), Gaps = 16/104 (15%)
Query 85 DLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTED 144
DLP EFD R E G++ P V++QG CGSCW+F A A+ +AT K V +S +
Sbjct 139 DLPTEFDWR-EQGAVTP----VKNQGMCGSCWSFSAIGALEGAHFLAT--KELVSLSEQQ 191
Query 145 LLTC---CD-----SCGFGCNGGYPQSAWEF-FKTKGIVTGGPY 179
L+ C CD SC GC+GG +A+E+ K G++ Y
Sbjct 192 LVDCDHECDPAQANSCDSGCSGGLMNNAFEYALKAGGLMKEEDY 235
> ath:AT5G45890 SAG12; SAG12 (SENESCENCE-ASSOCIATED GENE 12);
cysteine-type peptidase
Length=346
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 46/160 (28%), Positives = 77/160 (48%), Gaps = 22/160 (13%)
Query 29 MFQSHIHTINNMKT-----TWEAGENFGPHITSDYIRNLCGALK-------TPLSKKLPI 76
+F++++ I ++ + T++ N +T+D R++ K +K P
Sbjct 61 VFKNNVERIEHLNSIPAGRTFKLAVNQFADLTNDEFRSMYTGFKGVSALSSQSQTKMSPF 120
Query 77 KDLSKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKN 136
+ + LP+ D RK+ G++ P +++QG CG CWAF A A+ AT+ K
Sbjct 121 RYQNVSSGALPVSVDWRKK-GAVTP----IKNQGSCGCCWAFSAVAAIEG----ATQIKK 171
Query 137 QVRIS-TEDLLTCCDSCGFGCNGGYPQSAWEFFKTKGIVT 175
IS +E L CD+ FGC GG +A+E K G +T
Sbjct 172 GKLISLSEQQLVDCDTNDFGCEGGLMDTAFEHIKATGGLT 211
> ath:AT2G21430 cysteine proteinase A494, putative / thiol protease,
putative
Length=361
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 42/105 (40%), Positives = 58/105 (55%), Gaps = 16/105 (15%)
Query 84 HDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTE 143
+LP EFD R + G++ P V++QG CGSCW+F A+ +AT GK V +S +
Sbjct 130 QNLPEEFDWR-DRGAVTP----VKNQGSCGSCWSFSTTGALEGAHFLAT-GK-LVSLSEQ 182
Query 144 DLLTC---CD-----SCGFGCNGGYPQSAWEF-FKTKGIVTGGPY 179
L+ C CD SC GCNGG SA+E+ KT G++ Y
Sbjct 183 QLVDCDHECDPEEEGSCDSGCNGGLMNSAFEYTLKTGGLMREKDY 227
> ath:AT3G48340 cysteine-type endopeptidase/ cysteine-type peptidase
Length=296
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 46/140 (32%), Positives = 65/140 (46%), Gaps = 18/140 (12%)
Query 36 TINNMKTTWEAGENFGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEVHDLPIEFDARKE 95
TIN K + G N H R L G + ++LSK LP D RK+
Sbjct 24 TINEFKNAY-TGSNIKHH------RMLQGPKRGSKQFMYDHENLSK----LPSSVDWRKK 72
Query 96 WGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCDSCGFG 155
++ E+++QG+CGSCWAF A+ I T V +S ++L+ C G
Sbjct 73 -----GAVTEIKNQGKCGSCWAFSTVAAVEGINKIKT--NKLVSLSEQELVDCDTKQNEG 125
Query 156 CNGGYPQSAWEFFKTKGIVT 175
CNGG + A+EF K G +T
Sbjct 126 CNGGLMEIAFEFIKKNGGIT 145
> ath:AT1G06260 cysteine proteinase, putative
Length=343
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 43/154 (27%), Positives = 76/154 (49%), Gaps = 12/154 (7%)
Query 24 MPKNSMFQSHIHTI---NNMKTTWEAGEN-FGPHITSDYIRNLCGALKTPLSKKLPIKDL 79
M + ++QS++ I N++ ++ +N F S++ + G + L + +
Sbjct 61 MLRFGIYQSNVQLIDYINSLHLPFKLTDNRFADMTNSEFKAHFLGLNTSSLRLHKKQRPV 120
Query 80 SKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVR 139
++P D R + G++ P +R+QG+CG CWAF A A+ I T N V
Sbjct 121 CDPAGNVPDAVDWRTQ-GAVTP----IRNQGKCGGCWAFSAVAAIEGINKIKT--GNLVS 173
Query 140 ISTEDLLTC-CDSCGFGCNGGYPQSAWEFFKTKG 172
+S + L+ C + GC+GG ++A+EF KT G
Sbjct 174 LSEQQLIDCDVGTYNKGCSGGLMETAFEFIKTNG 207
> cel:F41E6.6 tag-196; Temporarily Assigned Gene name family member
(tag-196); K01373 cathepsin F [EC:3.4.22.41]
Length=477
Score = 56.6 bits (135), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 33/89 (37%), Positives = 49/89 (55%), Gaps = 8/89 (8%)
Query 78 DLSKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQ 137
D++ DLP FD R++ ++ +V++QG CGSCWAF + IA KN+
Sbjct 256 DVTINEEDLPESFDWREK-----GAVTQVKNQGNCGSCWAFSTTGNVEGAWFIA---KNK 307
Query 138 VRISTEDLLTCCDSCGFGCNGGYPQSAWE 166
+ +E L CDS GCNGG P +A++
Sbjct 308 LVSLSEQELVDCDSMDQGCNGGLPSNAYK 336
> ath:AT1G20850 XCP2; XCP2 (xylem cysteine peptidase 2); cysteine-type
peptidase/ peptidase
Length=356
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 41/157 (26%), Positives = 74/157 (47%), Gaps = 13/157 (8%)
Query 20 MVFTMPKNSMFQSHIHTINNMKTTWEAGENFGPHITSDYIRNLCGALKTPLSKKLPIKDL 79
+ F + K+++ HI N ++ G N ++ + + + LKT + ++ +
Sbjct 70 LRFEVFKDNL--KHIDETNKKGKSYWLGLNEFADLSHEEFKKMYLGLKTDIVRRDEERSY 127
Query 80 S----KEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGK 135
+ ++V +P D RK+ ++ EV++QG CGSCWAF A+ I T
Sbjct 128 AEFAYRDVEAVPKSVDWRKK-----GAVAEVKNQGSCGSCWAFSTVAAVEGINKIVT--G 180
Query 136 NQVRISTEDLLTCCDSCGFGCNGGYPQSAWEFFKTKG 172
N +S ++L+ C + GCNGG A+E+ G
Sbjct 181 NLTTLSEQELIDCDTTYNNGCNGGLMDYAFEYIVKNG 217
> ath:AT3G43960 cysteine proteinase, putative; K01376 [EC:3.4.22.-]
Length=376
Score = 55.8 bits (133), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 50/154 (32%), Positives = 77/154 (50%), Gaps = 10/154 (6%)
Query 22 FTMPKNSMFQSHIHTINNMKTTWEAGENFGPHITSDYIRN--LCGALKTPLSKKLPIKDL 79
F + K+++ + H ++ ++E G N +T+D + L G ++ + +
Sbjct 62 FKIFKDNLKRIEEHN-SDPNRSYERGLNKFSDLTADEFQASYLGGKMEKKSLSDVAERYQ 120
Query 80 SKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVR 139
KE LP E D R E G++ P V+ QGECGSCWAF AA + I T G+ V
Sbjct 121 YKEGDVLPDEVDWR-ERGAVVP---RVKRQGECGSCWAF-AATGAVEGINQITTGE-LVS 174
Query 140 ISTEDLLTC-CDSCGFGCNGGYPQSAWEFFKTKG 172
+S ++L+ C + FGC GG A+EF K G
Sbjct 175 LSEQELIDCDRGNDNFGCAGGGAVWAFEFIKENG 208
> ath:AT5G43060 cysteine proteinase, putative / thiol protease,
putative; K01376 [EC:3.4.22.-]
Length=463
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 47/156 (30%), Positives = 75/156 (48%), Gaps = 12/156 (7%)
Query 29 MFQSHIHTINNMKT---TWEAGENFGPHITSDYIRNL-CGALKTPLSKKLPIKDLSKEVH 84
+F+ ++ I+ T +++ G +T++ R++ GA T K + ++
Sbjct 77 IFKDNLRFIDEHNTKNLSYKLGLTRFADLTNEEYRSMYLGAKPTKRVLKTSDRYQARVGD 136
Query 85 DLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTED 144
LP D RKE ++ +V+DQG CGSCWAF A+ I T + + +S ++
Sbjct 137 ALPDSVDWRKE-----GAVADVKDQGSCGSCWAFSTIGAVEGINKIVT--GDLISLSEQE 189
Query 145 LLTCCDSCGFGCNGGYPQSAWEF-FKTKGIVTGGPY 179
L+ C S GCNGG A+EF K GI T Y
Sbjct 190 LVDCDTSYNQGCNGGLMDYAFEFIIKNGGIDTEADY 225
> mmu:13038 Ctsk, AI323530, MMS10-Q, Ms10q, catK; cathepsin K
(EC:3.4.22.38); K01371 cathepsin K [EC:3.4.22.38]
Length=329
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 38/132 (28%), Positives = 66/132 (50%), Gaps = 10/132 (7%)
Query 43 TWEAGENFGPHITSDYIRNLCGALKTPLSKKLPIKDLSKEVHD--LPIEFDARKEWGSIC 100
T+E N +TS+ + L+ P S+ L + +P D RK+ G +
Sbjct 70 TYELAMNHLGDMTSEEVVQKMTGLRIPPSRSYSNDTLYTPEWEGRVPDSIDYRKK-GYVT 128
Query 101 PSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCDSCGFGCNGGY 160
P V++QG+CGSCWAF +A A+ + + K + +S ++L+ C +GC GGY
Sbjct 129 P----VKNQGQCGSCWAFSSAGALEGQ--LKKKTGKLLALSPQNLVDCVTE-NYGCGGGY 181
Query 161 PQSAWEFFKTKG 172
+A+++ + G
Sbjct 182 MTTAFQYVQQNG 193
> ath:AT1G47128 RD21; RD21 (responsive to dehydration 21); cysteine-type
endopeptidase/ cysteine-type peptidase; K01376 [EC:3.4.22.-]
Length=462
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 46/161 (28%), Positives = 77/161 (47%), Gaps = 12/161 (7%)
Query 22 FTMPKNSMFQSHIHTINNMKTTWEAGENFGPHITSDYIRN--LCGALKTPLSKKLPIKDL 79
F + K+++ H N+ ++ G +T+D R+ L ++ ++ ++
Sbjct 73 FEIFKDNLRFVDEHNEKNL--SYRLGLTRFADLTNDEYRSKYLGAKMEKKGERRTSLRYE 130
Query 80 SKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVR 139
++ +LP D RK+ ++ EV+DQG CGSCWAF A+ I T + +
Sbjct 131 ARVGDELPESIDWRKK-----GAVAEVKDQGGCGSCWAFSTIGAVEGINQIVT--GDLIT 183
Query 140 ISTEDLLTCCDSCGFGCNGGYPQSAWEF-FKTKGIVTGGPY 179
+S ++L+ C S GCNGG A+EF K GI T Y
Sbjct 184 LSEQELVDCDTSYNEGCNGGLMDYAFEFIIKNGGIDTDKDY 224
> bbo:BBOV_IV007730 23.m06535; cysteine protease 2
Length=445
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 39/68 (57%), Gaps = 4/68 (5%)
Query 106 VRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCDSCGFGCNGGYPQSAW 165
V+DQG CGSCWAF A ++ + + K VR+S ++L++ C GCNGGY A
Sbjct 251 VKDQGMCGSCWAFAAVGSVE---SLLKRQKTDVRLSEQELVS-CQLGNQGCNGGYSDYAL 306
Query 166 EFFKTKGI 173
+ K GI
Sbjct 307 NYIKFNGI 314
> mmu:13032 Ctsc, AI047818, DPP1, DPPI; cathepsin C (EC:3.4.14.1);
K01275 cathepsin C [EC:3.4.14.1]
Length=462
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/131 (26%), Positives = 64/131 (48%), Gaps = 6/131 (4%)
Query 34 IHTINNMKTTWEAG--ENFGPHITSDYIRNLCGALKTPLSKKLPIKD-LSKEVHDLPIEF 90
+ IN ++ +W A + + D IR + + P K P+ D + +++ +LP +
Sbjct 175 VKAINTVQKSWTATAYKEYEKMSLRDLIRRSGHSQRIPRPKPAPMTDEIQQQILNLPESW 234
Query 91 DARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCD 150
D R G + VR+Q CGSC++F + + RI I T +S +++++C
Sbjct 235 DWRNVQG--VNYVSPVRNQESCGSCYSFASMGMLEARIRILTNNSQTPILSPQEVVSCSP 292
Query 151 SCGFGCNGGYP 161
GC+GG+P
Sbjct 293 YAQ-GCDGGFP 302
> mmu:26898 Ctsj, CATP, CatRLP, Ctsp, MGC117662; cathepsin J;
K09599 cathepsin P [EC:3.4.22.-]
Length=333
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 37/96 (38%), Positives = 51/96 (53%), Gaps = 9/96 (9%)
Query 86 LPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTEDL 145
LP D R+E G + P VR+QG+CGSCWAF AA A+ ++ T N +S ++L
Sbjct 113 LPDYKDWREE-GYVTP----VRNQGKCGSCWAFAAAGAIEGQMFWKT--GNLTPLSVQNL 165
Query 146 LTCCDSCGF-GCNGGYPQSAWEF-FKTKGIVTGGPY 179
L C + G GC G A+E+ K KG+ Y
Sbjct 166 LDCSKTVGNKGCQSGTAHQAFEYVLKNKGLEAEATY 201
> mmu:13040 Ctss; cathepsin S (EC:3.4.22.27); K01368 cathepsin
S [EC:3.4.22.27]
Length=340
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 43/156 (27%), Positives = 72/156 (46%), Gaps = 17/156 (10%)
Query 20 MVFTMPKNSMFQSHIHTINNMKTTWEAGENFGPHITSDYIRNLCGALKTPLSKKLPIKDL 79
+ F M N + +HT ++ G N +T++ I GAL+ P +
Sbjct 64 LKFIMIHNLEYSMGMHT-------YQVGMNDMGDMTNEEILCRMGALRIPRQSPKTVTFR 116
Query 80 SKEVHDLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVR 139
S LP D R++ + EV+ QG CG+CWAF A A+ ++ + T GK +
Sbjct 117 SYSNRTLPDTVDWREK-----GCVTEVKYQGSCGACWAFSAVGALEGQLKLKT-GK-LIS 169
Query 140 ISTEDLLTCCDSCGF---GCNGGYPQSAWEFFKTKG 172
+S ++L+ C + + GC GGY A+++ G
Sbjct 170 LSAQNLVDCSNEEKYGNKGCGGGYMTEAFQYIIDNG 205
> mmu:56092 Cts7, AA408230, AA408952, CTS1, Epcs24, Epcs71; cathepsin
7
Length=331
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 60/120 (50%), Gaps = 11/120 (9%)
Query 54 ITSDYIRNLCGALKTPLSKKLPIKDLSKEVHDLPIEFDARKEWGSICPSLLEVRDQGECG 113
+T + ++ L + PL K + K +P D RKE G + P VR QG CG
Sbjct 83 MTGEEMKMLTESSSYPLRNG---KHIQKRNPKIPPTLDWRKE-GYVTP----VRRQGSCG 134
Query 114 SCWAFGAAEAMTDRICIATKGKNQVRISTEDLLTCCDSCGF-GCNGGYPQSAWEFFKTKG 172
+CWAF + ++ T GK + +S ++L+ C S G GC+GG P A+++ K G
Sbjct 135 ACWAFSVTACIEGQLFKKT-GK-LIPLSVQNLMDCSVSYGTKGCDGGRPYDAFQYVKNNG 192
> tpv:TP03_0283 cysteine proteinase (EC:3.4.22.-); K01376 [EC:3.4.22.-]
Length=441
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 40/126 (31%), Positives = 61/126 (48%), Gaps = 27/126 (21%)
Query 68 TPLSKKLPIKDLS----------------KEVHDLPIEFDARKEWGSICPSLLEVRDQGE 111
T LSK L K +S +E+ D+ + +W + ++ V+DQG+
Sbjct 189 TSLSKHLEFKKMSYKNPIYISKLKEAKGVEEIKDISLVTGENLDW-TRTDTVSPVKDQGD 247
Query 112 -CGSCWAF---GAAEAMTDRICIATKGKNQVRISTEDLLTCCDSCGFGCNGGYPQSAWEF 167
CGSCWAF G+ E++ R+ KN+ +E L CD GC+GG P +A E+
Sbjct 248 HCGSCWAFSSIGSVESLY-RLY-----KNKSYFLSEQELVNCDKSSMGCSGGLPITAMEY 301
Query 168 FKTKGI 173
+KGI
Sbjct 302 IHSKGI 307
> mmu:70202 Ctsll3, 2310051M13Rik; cathepsin L-like 3 (EC:3.4.22.15)
Length=331
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 35/89 (39%), Positives = 50/89 (56%), Gaps = 8/89 (8%)
Query 85 DLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTED 144
D+P D R G + P V+DQG CGSCWAF A ++ ++ T GK V +S ++
Sbjct 114 DVPKSVDWRDH-GYVTP----VKDQGSCGSCWAFSAVGSLEGQMFRKT-GK-LVPLSVQN 166
Query 145 LLTCCDSCG-FGCNGGYPQSAWEFFKTKG 172
L+ C S G GC+GG P A+++ K G
Sbjct 167 LVDCSWSQGNQGCDGGLPDLAFQYVKDNG 195
> mmu:218275 MGC60730; cDNA sequence BC051665
Length=330
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 38/97 (39%), Positives = 54/97 (55%), Gaps = 9/97 (9%)
Query 85 DLPIEFDARKEWGSICPSLLEVRDQGECGSCWAFGAAEAMTDRICIATKGKNQVRISTED 144
D+P D R G + P V+DQG CGSCWAF A ++ +I T GK V +S ++
Sbjct 113 DVPKSVDWRDH-GYVTP----VKDQGHCGSCWAFSAVGSLEGQIFRKT-GK-LVPLSEQN 165
Query 145 LLTCCDSCG-FGCNGGYPQSAWEFFK-TKGIVTGGPY 179
L+ C S G GCNGG + A+++ K +G+ T Y
Sbjct 166 LMDCSWSYGNVGCNGGLMELAFQYVKENRGLDTRESY 202
Lambda K H
0.321 0.137 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4924747408
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40