bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7474_orf1
Length=182
Score E
Sequences producing significant alignments: (Bits) Value
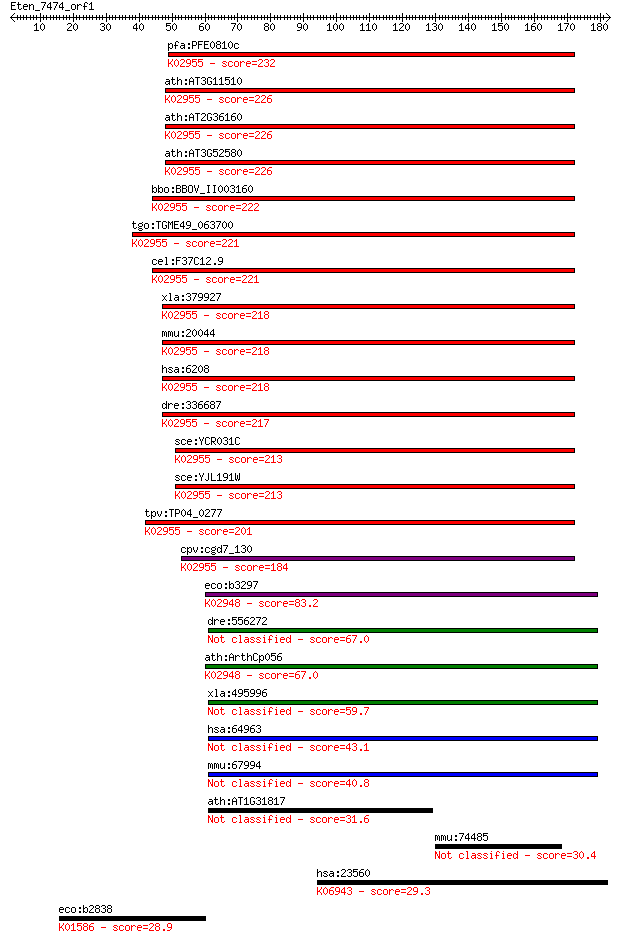
pfa:PFE0810c 40S ribosomal protein S14, putative; K02955 small... 232 7e-61
ath:AT3G11510 40S ribosomal protein S14 (RPS14B); K02955 small... 226 3e-59
ath:AT2G36160 40S ribosomal protein S14 (RPS14A); K02955 small... 226 4e-59
ath:AT3G52580 40S ribosomal protein S14 (RPS14C); K02955 small... 226 4e-59
bbo:BBOV_II003160 18.m06264; 40S ribosomal subunit protein S14... 222 4e-58
tgo:TGME49_063700 40s ribosomal protein S14, putative ; K02955... 221 1e-57
cel:F37C12.9 rps-14; Ribosomal Protein, Small subunit family m... 221 1e-57
xla:379927 rps14, MGC130808, MGC53662; ribosomal protein S14; ... 218 1e-56
mmu:20044 Rps14, 2600014J02Rik, AL023078, MGC86125; ribosomal ... 218 1e-56
hsa:6208 RPS14, EMTB; ribosomal protein S14; K02955 small subu... 218 1e-56
dre:336687 rps14, fa92e08, wu:fa92e08, zgc:73215; ribosomal pr... 217 2e-56
sce:YCR031C RPS14A, CRY1, RPL59; Rps14ap; K02955 small subunit... 213 3e-55
sce:YJL191W RPS14B, CRY2; Rps14bp; K02955 small subunit riboso... 213 3e-55
tpv:TP04_0277 40S ribosomal protein S14; K02955 small subunit ... 201 2e-51
cpv:cgd7_130 40S ribosomal protein S14 ; K02955 small subunit ... 184 1e-46
eco:b3297 rpsK, ECK3284, JW3259; 30S ribosomal subunit protein... 83.2 5e-16
dre:556272 MGC162401, MGC198257, MGC198259, zgc:162401; si:dke... 67.0 3e-11
ath:ArthCp056 rps11; 30S ribosomal protein S11; K02948 small s... 67.0 4e-11
xla:495996 mrps11; mitochondrial ribosomal protein S11 59.7 5e-09
hsa:64963 MRPS11, FLJ22512, FLJ23406, HCC-2; mitochondrial rib... 43.1 5e-04
mmu:67994 Mrps11, 0710005I03Rik, C79873; mitochondrial ribosom... 40.8 0.003
ath:AT1G31817 NFD3; NFD3 (NUCLEAR FUSION DEFECTIVE 3); structu... 31.6 1.6
mmu:74485 4933430H15Rik, Lrrc71; RIKEN cDNA 4933430H15 gene 30.4 3.6
hsa:23560 GTPBP4, CRFG, FLJ10686, FLJ10690, FLJ39774, NGB, NOG... 29.3 7.1
eco:b2838 lysA, ECK2836, JW2806; diaminopimelate decarboxylase... 28.9 9.4
> pfa:PFE0810c 40S ribosomal protein S14, putative; K02955 small
subunit ribosomal protein S14e
Length=151
Score = 232 bits (591), Expect = 7e-61, Method: Compositional matrix adjust.
Identities = 109/123 (88%), Positives = 119/123 (96%), Gaps = 0/123 (0%)
Query 49 GPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMA 108
GPQ +E ELVFGVAHI+ASFNDTFIHVTDLSGRETLVR+TGGMKVKADRDESSPYAAMMA
Sbjct 18 GPQPKEGELVFGVAHIFASFNDTFIHVTDLSGRETLVRITGGMKVKADRDESSPYAAMMA 77
Query 109 AGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPT 168
A DVAAR KELG+TA+H+++RA+GGTKSKTPGPGAQSALRALARSGL+IGRIEDVTPIPT
Sbjct 78 AQDVAARLKELGVTAIHIKLRASGGTKSKTPGPGAQSALRALARSGLKIGRIEDVTPIPT 137
Query 169 DST 171
DST
Sbjct 138 DST 140
> ath:AT3G11510 40S ribosomal protein S14 (RPS14B); K02955 small
subunit ribosomal protein S14e
Length=150
Score = 226 bits (576), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 107/124 (86%), Positives = 116/124 (93%), Gaps = 0/124 (0%)
Query 48 LGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMM 107
LGP RE E VFGV HI+ASFNDTFIHVTDLSGRETLVR+TGGMKVKADRDESSPYAAM+
Sbjct 16 LGPSVREGEQVFGVVHIFASFNDTFIHVTDLSGRETLVRITGGMKVKADRDESSPYAAML 75
Query 108 AAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIP 167
AA DVA RCKELGITA+HV++RATGG K+KTPGPGAQSALRALARSG++IGRIEDVTPIP
Sbjct 76 AAQDVAQRCKELGITAMHVKLRATGGNKTKTPGPGAQSALRALARSGMKIGRIEDVTPIP 135
Query 168 TDST 171
TDST
Sbjct 136 TDST 139
> ath:AT2G36160 40S ribosomal protein S14 (RPS14A); K02955 small
subunit ribosomal protein S14e
Length=150
Score = 226 bits (576), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 107/124 (86%), Positives = 116/124 (93%), Gaps = 0/124 (0%)
Query 48 LGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMM 107
LGP RE E VFGV HI+ASFNDTFIHVTDLSGRETLVR+TGGMKVKADRDESSPYAAM+
Sbjct 16 LGPSVREGEQVFGVVHIFASFNDTFIHVTDLSGRETLVRITGGMKVKADRDESSPYAAML 75
Query 108 AAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIP 167
AA DVA RCKELGITA+HV++RATGG K+KTPGPGAQSALRALARSG++IGRIEDVTPIP
Sbjct 76 AAQDVAQRCKELGITAMHVKLRATGGNKTKTPGPGAQSALRALARSGMKIGRIEDVTPIP 135
Query 168 TDST 171
TDST
Sbjct 136 TDST 139
> ath:AT3G52580 40S ribosomal protein S14 (RPS14C); K02955 small
subunit ribosomal protein S14e
Length=150
Score = 226 bits (575), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 106/124 (85%), Positives = 116/124 (93%), Gaps = 0/124 (0%)
Query 48 LGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMM 107
LGP RE E VFGV H++ASFNDTFIHVTDLSGRETLVR+TGGMKVKADRDESSPYAAM+
Sbjct 16 LGPAVREGEQVFGVVHVFASFNDTFIHVTDLSGRETLVRITGGMKVKADRDESSPYAAML 75
Query 108 AAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIP 167
AA DVA RCKELGITA+HV++RATGG K+KTPGPGAQSALRALARSG++IGRIEDVTPIP
Sbjct 76 AAQDVAQRCKELGITAIHVKLRATGGNKTKTPGPGAQSALRALARSGMKIGRIEDVTPIP 135
Query 168 TDST 171
TDST
Sbjct 136 TDST 139
> bbo:BBOV_II003160 18.m06264; 40S ribosomal subunit protein S14;
K02955 small subunit ribosomal protein S14e
Length=168
Score = 222 bits (566), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 110/128 (85%), Positives = 118/128 (92%), Gaps = 1/128 (0%)
Query 44 AAAALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPY 103
A LGPQ + VFGVAHI+ASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPY
Sbjct 31 ATTPLGPQQKGGH-VFGVAHIFASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPY 89
Query 104 AAMMAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDV 163
AAMMAA DVAAR KELGITAVH+++RATGGT+SKTPGPGAQSALR+LARSGL+IGRIEDV
Sbjct 90 AAMMAAQDVAARLKELGITAVHIKLRATGGTRSKTPGPGAQSALRSLARSGLKIGRIEDV 149
Query 164 TPIPTDST 171
TPIPTDST
Sbjct 150 TPIPTDST 157
> tgo:TGME49_063700 40s ribosomal protein S14, putative ; K02955
small subunit ribosomal protein S14e
Length=156
Score = 221 bits (563), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 119/134 (88%), Positives = 125/134 (93%), Gaps = 0/134 (0%)
Query 38 GELGAAAAAALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADR 97
GE G A + LGP TR+ E+VFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADR
Sbjct 12 GEEGEAQGSGLGPATRDGEIVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADR 71
Query 98 DESSPYAAMMAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRI 157
DESSPYAAMMAA DVAARCKELGI+AVHV+VRA GGT+SKTPGPGAQSALRALARSGLRI
Sbjct 72 DESSPYAAMMAAADVAARCKELGISAVHVKVRAAGGTRSKTPGPGAQSALRALARSGLRI 131
Query 158 GRIEDVTPIPTDST 171
GRIEDVTPIPTDST
Sbjct 132 GRIEDVTPIPTDST 145
> cel:F37C12.9 rps-14; Ribosomal Protein, Small subunit family
member (rps-14); K02955 small subunit ribosomal protein S14e
Length=152
Score = 221 bits (562), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 100/128 (78%), Positives = 120/128 (93%), Gaps = 0/128 (0%)
Query 44 AAAALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPY 103
A +LGPQ +E EL+FGVAHI+ASFNDTF+H+TD+SGRET+VRVTGGMKVKADRDESSPY
Sbjct 14 AVVSLGPQAKEGELIFGVAHIFASFNDTFVHITDISGRETIVRVTGGMKVKADRDESSPY 73
Query 104 AAMMAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDV 163
AAM+AA DVA RCK+LGI A+H+++RATGGT++KTPGPGAQSALRALAR+G++IGRIEDV
Sbjct 74 AAMLAAQDVADRCKQLGINALHIKLRATGGTRTKTPGPGAQSALRALARAGMKIGRIEDV 133
Query 164 TPIPTDST 171
TPIP+D T
Sbjct 134 TPIPSDCT 141
> xla:379927 rps14, MGC130808, MGC53662; ribosomal protein S14;
K02955 small subunit ribosomal protein S14e
Length=151
Score = 218 bits (554), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 101/125 (80%), Positives = 116/125 (92%), Gaps = 0/125 (0%)
Query 47 ALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAM 106
+LGPQ E E VFGV HI+ASFNDTF+HVTDLSG+ET+ RVTGGMKVKADRDESSPYAAM
Sbjct 16 SLGPQVAEGENVFGVCHIFASFNDTFVHVTDLSGKETICRVTGGMKVKADRDESSPYAAM 75
Query 107 MAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPI 166
+AA DVA RCKELGITA+H+++RATGG ++KTPGPGAQSALRALARSG++IGRIEDVTPI
Sbjct 76 LAAQDVAQRCKELGITALHIKLRATGGNRTKTPGPGAQSALRALARSGMKIGRIEDVTPI 135
Query 167 PTDST 171
P+DST
Sbjct 136 PSDST 140
> mmu:20044 Rps14, 2600014J02Rik, AL023078, MGC86125; ribosomal
protein S14; K02955 small subunit ribosomal protein S14e
Length=151
Score = 218 bits (554), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 101/125 (80%), Positives = 116/125 (92%), Gaps = 0/125 (0%)
Query 47 ALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAM 106
+LGPQ E E VFGV HI+ASFNDTF+HVTDLSG+ET+ RVTGGMKVKADRDESSPYAAM
Sbjct 16 SLGPQVAEGENVFGVCHIFASFNDTFVHVTDLSGKETICRVTGGMKVKADRDESSPYAAM 75
Query 107 MAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPI 166
+AA DVA RCKELGITA+H+++RATGG ++KTPGPGAQSALRALARSG++IGRIEDVTPI
Sbjct 76 LAAQDVAQRCKELGITALHIKLRATGGNRTKTPGPGAQSALRALARSGMKIGRIEDVTPI 135
Query 167 PTDST 171
P+DST
Sbjct 136 PSDST 140
> hsa:6208 RPS14, EMTB; ribosomal protein S14; K02955 small subunit
ribosomal protein S14e
Length=151
Score = 218 bits (554), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 101/125 (80%), Positives = 116/125 (92%), Gaps = 0/125 (0%)
Query 47 ALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAM 106
+LGPQ E E VFGV HI+ASFNDTF+HVTDLSG+ET+ RVTGGMKVKADRDESSPYAAM
Sbjct 16 SLGPQVAEGENVFGVCHIFASFNDTFVHVTDLSGKETICRVTGGMKVKADRDESSPYAAM 75
Query 107 MAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPI 166
+AA DVA RCKELGITA+H+++RATGG ++KTPGPGAQSALRALARSG++IGRIEDVTPI
Sbjct 76 LAAQDVAQRCKELGITALHIKLRATGGNRTKTPGPGAQSALRALARSGMKIGRIEDVTPI 135
Query 167 PTDST 171
P+DST
Sbjct 136 PSDST 140
> dre:336687 rps14, fa92e08, wu:fa92e08, zgc:73215; ribosomal
protein S14; K02955 small subunit ribosomal protein S14e
Length=151
Score = 217 bits (552), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 100/125 (80%), Positives = 116/125 (92%), Gaps = 0/125 (0%)
Query 47 ALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAM 106
+LGPQ E E VFGV HI+ASFNDTF+HVTDLSG+ET+ RVTGGMKVKADRDESSPYAAM
Sbjct 16 SLGPQVAEGENVFGVCHIFASFNDTFVHVTDLSGKETICRVTGGMKVKADRDESSPYAAM 75
Query 107 MAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPI 166
+AA DVA +CKELGITA+H+++RATGG ++KTPGPGAQSALRALARSG++IGRIEDVTPI
Sbjct 76 LAAQDVAQKCKELGITALHIKLRATGGNRTKTPGPGAQSALRALARSGMKIGRIEDVTPI 135
Query 167 PTDST 171
P+DST
Sbjct 136 PSDST 140
> sce:YCR031C RPS14A, CRY1, RPL59; Rps14ap; K02955 small subunit
ribosomal protein S14e
Length=137
Score = 213 bits (542), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 99/121 (81%), Positives = 114/121 (94%), Gaps = 0/121 (0%)
Query 51 QTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAG 110
Q R++ VFGVA IYASFNDTF+HVTDLSG+ET+ RVTGGMKVKADRDESSPYAAM+AA
Sbjct 6 QARDNSQVFGVARIYASFNDTFVHVTDLSGKETIARVTGGMKVKADRDESSPYAAMLAAQ 65
Query 111 DVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDS 170
DVAA+CKE+GITAVHV++RATGGT++KTPGPG Q+ALRALARSGLRIGRIEDVTP+P+DS
Sbjct 66 DVAAKCKEVGITAVHVKIRATGGTRTKTPGPGGQAALRALARSGLRIGRIEDVTPVPSDS 125
Query 171 T 171
T
Sbjct 126 T 126
> sce:YJL191W RPS14B, CRY2; Rps14bp; K02955 small subunit ribosomal
protein S14e
Length=138
Score = 213 bits (542), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 99/121 (81%), Positives = 114/121 (94%), Gaps = 0/121 (0%)
Query 51 QTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAG 110
Q R++ VFGVA IYASFNDTF+HVTDLSG+ET+ RVTGGMKVKADRDESSPYAAM+AA
Sbjct 7 QARDNSQVFGVARIYASFNDTFVHVTDLSGKETIARVTGGMKVKADRDESSPYAAMLAAQ 66
Query 111 DVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDS 170
DVAA+CKE+GITAVHV++RATGGT++KTPGPG Q+ALRALARSGLRIGRIEDVTP+P+DS
Sbjct 67 DVAAKCKEVGITAVHVKIRATGGTRTKTPGPGGQAALRALARSGLRIGRIEDVTPVPSDS 126
Query 171 T 171
T
Sbjct 127 T 127
> tpv:TP04_0277 40S ribosomal protein S14; K02955 small subunit
ribosomal protein S14e
Length=151
Score = 201 bits (510), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 109/130 (83%), Positives = 118/130 (90%), Gaps = 1/130 (0%)
Query 42 AAAAAALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESS 101
A LGPQ + E VFGVAH++ASFNDTFIH+TDLSGRETLVRVTGGMKVKADRDESS
Sbjct 12 AEVPTTLGPQVK-GEHVFGVAHVFASFNDTFIHITDLSGRETLVRVTGGMKVKADRDESS 70
Query 102 PYAAMMAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIE 161
PYAAMMAA D AAR KELGITAVHV++RATGGT+SKTPGPGAQSALR+LARSGL+IGRIE
Sbjct 71 PYAAMMAAQDAAARAKELGITAVHVKLRATGGTRSKTPGPGAQSALRSLARSGLKIGRIE 130
Query 162 DVTPIPTDST 171
DVTPIPTDST
Sbjct 131 DVTPIPTDST 140
> cpv:cgd7_130 40S ribosomal protein S14 ; K02955 small subunit
ribosomal protein S14e
Length=131
Score = 184 bits (468), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 96/119 (80%), Positives = 108/119 (90%), Gaps = 0/119 (0%)
Query 53 REDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDV 112
RE + F VAHI+ASFNDTFIHVTDLSGRET+VRVTGGMKVKADRDE+SPYAAMMAA DV
Sbjct 2 REGQTNFAVAHIFASFNDTFIHVTDLSGRETIVRVTGGMKVKADRDENSPYAAMMAAVDV 61
Query 113 AARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDST 171
A +CKE GI A+H+++RA GG KSKT GPGAQSALRALAR+G++IGRIEDVTPIPTDST
Sbjct 62 AQKCKEFGIHALHIKLRAVGGVKSKTMGPGAQSALRALARAGMKIGRIEDVTPIPTDST 120
> eco:b3297 rpsK, ECK3284, JW3259; 30S ribosomal subunit protein
S11; K02948 small subunit ribosomal protein S11
Length=129
Score = 83.2 bits (204), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 51/119 (42%), Positives = 65/119 (54%), Gaps = 9/119 (7%)
Query 60 GVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKEL 119
GVAHI+ASFN+T + +TD G GG + R +S+P+AA +AA A KE
Sbjct 19 GVAHIHASFNNTIVTITDRQGNALGWATAGGSGFRGSR-KSTPFAAQVAAERCADAVKEY 77
Query 120 GITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDSTRRKSGRR 178
GI + V V K PGPG +S +RAL +G RI I DVTPIP + R RR
Sbjct 78 GIKNLEVMV--------KGPGPGRESTIRALNAAGFRITNITDVTPIPHNGCRPPKKRR 128
> dre:556272 MGC162401, MGC198257, MGC198259, zgc:162401; si:dkey-265m8.2
Length=199
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 64/118 (54%), Gaps = 9/118 (7%)
Query 61 VAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKELG 120
+AH+ A++N+T I VTD +G + +VR + G + + +S+P AA A AA+ + G
Sbjct 90 IAHVKATYNNTHIQVTDCAG-QYMVRTSCGTEGFKNVKKSTPIAAQTAGISAAAKARAKG 148
Query 121 ITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDSTRRKSGRR 178
+T V V V+ G PG SA++ L GL + I D TP+P + R + RR
Sbjct 149 VTYVRVLVKGLG--------PGRFSAIKGLTMGGLEVVSITDNTPVPHNGCRPRKARR 198
> ath:ArthCp056 rps11; 30S ribosomal protein S11; K02948 small
subunit ribosomal protein S11
Length=138
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 43/119 (36%), Positives = 61/119 (51%), Gaps = 9/119 (7%)
Query 60 GVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKEL 119
GV H+ ASFN+T + VTD+ GR G + R +P+AA AAG+ +
Sbjct 28 GVIHVQASFNNTIVTVTDVRGRVISWSSAGTCGFRGTR-RGTPFAAQTAAGNAIRAVVDQ 86
Query 120 GITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDSTRRKSGRR 178
G+ VR+ K PG G +ALRA+ RSG+ + + DVTP+P + R RR
Sbjct 87 GMQRAEVRI--------KGPGLGRDAALRAIRRSGILLSFVRDVTPMPHNGCRPPKKRR 137
> xla:495996 mrps11; mitochondrial ribosomal protein S11
Length=195
Score = 59.7 bits (143), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 61/118 (51%), Gaps = 9/118 (7%)
Query 61 VAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKELG 120
+AH+ A++N+T I VT ++LVR + G + + +S+ AA A ++A K +G
Sbjct 86 IAHVKATYNNTHIQVTSYEN-QSLVRTSCGTEGFKNAKKSTSVAAQTAG--ISAAAKAIG 142
Query 121 ITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDSTRRKSGRR 178
HVRV G GPG SA++ L GL + I D TPIP + R + RR
Sbjct 143 KGVSHVRVVVKG------LGPGRLSAIKGLTMGGLEVISITDDTPIPHNGCRPRKARR 194
> hsa:64963 MRPS11, FLJ22512, FLJ23406, HCC-2; mitochondrial ribosomal
protein S11
Length=194
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 57/118 (48%), Gaps = 9/118 (7%)
Query 61 VAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKELG 120
+AHI AS N+T I V S E L + G + + + + AA A AAR K+ G
Sbjct 85 IAHIKASHNNTQIQVVSASN-EPLAFASCGTEGFRNAKKGTGIAAQTAGIAAAARAKQKG 143
Query 121 ITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDSTRRKSGRR 178
+ +H+RV G GPG SA+ L GL + I D TPIP + R + R+
Sbjct 144 V--IHIRVVVKGL------GPGRLSAMHGLIMGGLEVISITDNTPIPHNGCRPRKARK 193
> mmu:67994 Mrps11, 0710005I03Rik, C79873; mitochondrial ribosomal
protein S11
Length=191
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 38/118 (32%), Positives = 58/118 (49%), Gaps = 9/118 (7%)
Query 61 VAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKELG 120
+AHI A++N+T I V + +L R + G + + + + AA A AA+ G
Sbjct 82 IAHIKATYNNTQIQVVSATN-ASLARASCGTEGFRNAKKGTGIAAQTAGIAAAAKATGKG 140
Query 121 ITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDSTRRKSGRR 178
+T H+RV G GPG SA++ L GL + I D TP+P + R + RR
Sbjct 141 VT--HIRVVVKGM------GPGRWSAIKGLTMGGLEVISITDNTPVPHNGCRPRKARR 190
> ath:AT1G31817 NFD3; NFD3 (NUCLEAR FUSION DEFECTIVE 3); structural
constituent of ribosome
Length=314
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 35/69 (50%), Gaps = 2/69 (2%)
Query 61 VAHIYASFNDTFIHVTDLSGRETLVRVTGGM-KVKADRDESSPYAAMMAAGDVAARCKEL 119
+ HI N+TF+ VTD G +G + +K R ++ Y A A ++ R K +
Sbjct 195 IIHIKMLRNNTFVTVTDSKGNVKCKATSGSLPDLKGGRKMTN-YTADATAENIGRRAKAM 253
Query 120 GITAVHVRV 128
G+ +V V+V
Sbjct 254 GLKSVVVKV 262
> mmu:74485 4933430H15Rik, Lrrc71; RIKEN cDNA 4933430H15 gene
Length=558
Score = 30.4 bits (67), Expect = 3.6, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 130 ATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIP 167
+T GT +TP PGAQ + A+ + G R + + + +P
Sbjct 6 STTGTSPRTPRPGAQKSSGAVTKKGDRAAKDKTASTLP 43
> hsa:23560 GTPBP4, CRFG, FLJ10686, FLJ10690, FLJ39774, NGB, NOG1;
GTP binding protein 4; K06943 nucleolar GTP-binding protein
Length=634
Score = 29.3 bits (64), Expect = 7.1, Method: Composition-based stats.
Identities = 22/89 (24%), Positives = 44/89 (49%), Gaps = 7/89 (7%)
Query 94 KADRDESSPYAAMMAAGDVAARCKEL-GITAVHVRVRATGGTKSKTPGPGAQSALRALAR 152
K R++S+P +++ +G + +++ G+ V + K+KT AQ + L +
Sbjct 552 KRKREDSAPPSSVARSGSCSRTPRDVSGLRDVKMV------KKAKTMMKNAQKKMNRLGK 605
Query 153 SGLRIGRIEDVTPIPTDSTRRKSGRRGRR 181
G + D+ P S +RK+G++ RR
Sbjct 606 KGEADRHVFDMKPKHLLSGKRKAGKKDRR 634
> eco:b2838 lysA, ECK2836, JW2806; diaminopimelate decarboxylase,
PLP-binding (EC:4.1.1.20); K01586 diaminopimelate decarboxylase
[EC:4.1.1.20]
Length=420
Score = 28.9 bits (63), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 24/47 (51%), Gaps = 3/47 (6%)
Query 16 RKYCGPLAGLRV---AAVLERSCSQGELGAAAAAALGPQTREDELVF 59
+K C + LR+ V S S GE+ A AA PQT D++VF
Sbjct 53 QKACSNIHILRLMREQGVKVDSVSLGEIERALAAGYNPQTHPDDIVF 99
Lambda K H
0.319 0.133 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4912245712
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40