bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7453_orf1
Length=106
Score E
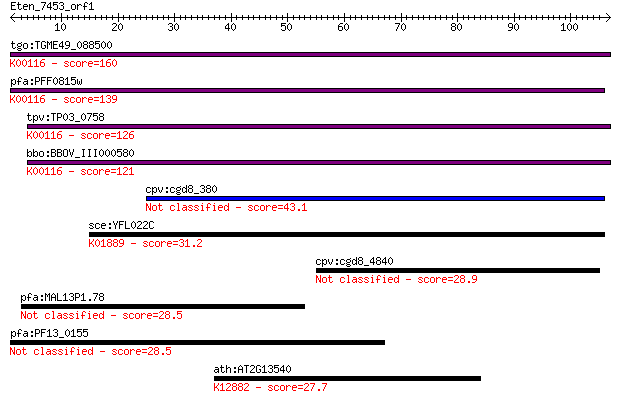
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_088500 malate:quinone oxidoreductase, putative (EC:... 160 1e-39
pfa:PFF0815w malate:quinone oxidoreductase, putative (EC:1.1.9... 139 2e-33
tpv:TP03_0758 malate:quinone oxidoreductase (EC:1.1.99.16); K0... 126 1e-29
bbo:BBOV_III000580 17.m07076; malate:quinone oxidoreductase (E... 121 5e-28
cpv:cgd8_380 possible oxidase or dehydrogenase 43.1 2e-04
sce:YFL022C FRS2; Frs2p (EC:6.1.1.20); K01889 phenylalanyl-tRN... 31.2 0.84
cpv:cgd8_4840 hypothetical protein 28.9 3.9
pfa:MAL13P1.78 conserved Plasmodium protein, unknown function 28.5 5.2
pfa:PF13_0155 conserved Plasmodium protein, unknown function 28.5 5.4
ath:AT2G13540 ABH1; ABH1 (ABA HYPERSENSITIVE 1); RNA cap bindi... 27.7 9.1
> tgo:TGME49_088500 malate:quinone oxidoreductase, putative (EC:1.1.99.16);
K00116 malate dehydrogenase (quinone) [EC:1.1.5.4]
Length=550
Score = 160 bits (405), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 75/106 (70%), Positives = 88/106 (83%), Gaps = 0/106 (0%)
Query 1 LAHVYVQLLNTAYMRAYVFKNFLFEVPALNLHLFAKDLRKIIPSIQVEDLTYAKGFGGIR 60
L VY+ LL T++MR YV +NFLFEVP LN LFA D++KI+PSI+ EDLTYA+G+GG+R
Sbjct 417 LVKVYLDLLGTSHMRNYVLRNFLFEVPKLNTVLFANDVKKIVPSIKPEDLTYAQGYGGVR 476
Query 61 PQLINKTEKKLLLGEGKIVTGENIIFNITPSPGGTTCLGTAETDMR 106
PQLI+K +KKLLLGEGKI G IIFNITPSPGGTTCLG AE DMR
Sbjct 477 PQLIDKVQKKLLLGEGKINPGTGIIFNITPSPGGTTCLGNAELDMR 522
> pfa:PFF0815w malate:quinone oxidoreductase, putative (EC:1.1.99.16);
K00116 malate dehydrogenase (quinone) [EC:1.1.5.4]
Length=521
Score = 139 bits (350), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 66/105 (62%), Positives = 78/105 (74%), Gaps = 0/105 (0%)
Query 1 LAHVYVQLLNTAYMRAYVFKNFLFEVPALNLHLFAKDLRKIIPSIQVEDLTYAKGFGGIR 60
L VY L M YV +N LFE+P LN +LF KD++KIIPS+ ++DLTY G+GG+R
Sbjct 387 LFQVYYNLFKDMTMLKYVARNVLFEIPVLNKYLFLKDVKKIIPSLTIKDLTYCVGYGGVR 446
Query 61 PQLINKTEKKLLLGEGKIVTGENIIFNITPSPGGTTCLGTAETDM 105
PQLINK KKL+LGEGKI G+NIIFNITPSPG TTCLG E DM
Sbjct 447 PQLINKKSKKLILGEGKIDPGKNIIFNITPSPGATTCLGNGEFDM 491
> tpv:TP03_0758 malate:quinone oxidoreductase (EC:1.1.99.16);
K00116 malate dehydrogenase (quinone) [EC:1.1.5.4]
Length=452
Score = 126 bits (317), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 56/103 (54%), Positives = 74/103 (71%), Gaps = 0/103 (0%)
Query 4 VYVQLLNTAYMRAYVFKNFLFEVPALNLHLFAKDLRKIIPSIQVEDLTYAKGFGGIRPQL 63
VY L + +R YV KNFLFEVP L+ LF +D RKI+PS+ D+ Y++G+GG+RPQL
Sbjct 322 VYFDLFKQSTIRNYVLKNFLFEVPLLSRWLFLRDARKIVPSLNYSDIVYSQGYGGVRPQL 381
Query 64 INKTEKKLLLGEGKIVTGENIIFNITPSPGGTTCLGTAETDMR 106
++K +KLL GE KI +G+ +IFNI+PSPG TTCL A D R
Sbjct 382 VDKKNRKLLFGESKIRSGDGLIFNISPSPGATTCLSNAVGDAR 424
> bbo:BBOV_III000580 17.m07076; malate:quinone oxidoreductase
(EC:1.1.99.16); K00116 malate dehydrogenase (quinone) [EC:1.1.5.4]
Length=490
Score = 121 bits (304), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 54/103 (52%), Positives = 74/103 (71%), Gaps = 0/103 (0%)
Query 4 VYVQLLNTAYMRAYVFKNFLFEVPALNLHLFAKDLRKIIPSIQVEDLTYAKGFGGIRPQL 63
VY+ LL + Y+F+N LFEVP L+ LF KD RKI+P++ ++DL Y+ G+GG+RPQL
Sbjct 362 VYLDLLKDTTICKYMFRNMLFEVPGLSRLLFLKDARKIVPTLTLKDLRYSVGYGGVRPQL 421
Query 64 INKTEKKLLLGEGKIVTGENIIFNITPSPGGTTCLGTAETDMR 106
++K KKLLLG+ K+ + I+NIT SPG TTC+ AE DMR
Sbjct 422 VDKVNKKLLLGQSKLTNNDGCIYNITASPGATTCISNAEIDMR 464
> cpv:cgd8_380 possible oxidase or dehydrogenase
Length=526
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 29/84 (34%), Positives = 45/84 (53%), Gaps = 3/84 (3%)
Query 25 EVPALNLHLFAKDLRKIIPSIQVEDLT-YAKGFGGIRPQLINKTEKK--LLLGEGKIVTG 81
++P L + RKIIPS+ + L + F QL++ + +K ++L KI T
Sbjct 409 KIPKLVSEQIIQGSRKIIPSLNLSQLKDKQQNFDISINQLLDVSGEKNQVILDRSKIHTD 468
Query 82 ENIIFNITPSPGGTTCLGTAETDM 105
EN+I NI+ GTTC+ +A DM
Sbjct 469 ENLILNISQPNSGTTCIKSATDDM 492
> sce:YFL022C FRS2; Frs2p (EC:6.1.1.20); K01889 phenylalanyl-tRNA
synthetase alpha chain [EC:6.1.1.20]
Length=503
Score = 31.2 bits (69), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 44/98 (44%), Gaps = 12/98 (12%)
Query 15 RAYVFKNFLFEVPALN-LHLFAK----DLRKIIPSIQ--VEDLTYAKGFGGIRPQLINKT 67
+A FKN A N L L AK DL ++ Q + + I +++N
Sbjct 103 QARAFKNGWIAKNASNELELSAKLQNTDLNELTDETQSILAQIKNNSHLDSIDAKILNDL 162
Query 68 EKKLLLGEGKIVTGENIIFNITPSPGGTTCLGTAETDM 105
+K+ L+ +GKI FN+T P +T L ETD+
Sbjct 163 KKRKLIAQGKITD-----FNVTKGPEFSTDLTKLETDL 195
> cpv:cgd8_4840 hypothetical protein
Length=2409
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 22/50 (44%), Gaps = 0/50 (0%)
Query 55 GFGGIRPQLINKTEKKLLLGEGKIVTGENIIFNITPSPGGTTCLGTAETD 104
GF G +N+ LLG G IV G N IT SP T+ + D
Sbjct 1916 GFVGGLANPLNQMIMNTLLGSGGIVGGSNYTAGITTSPNPTSVNNSTTKD 1965
> pfa:MAL13P1.78 conserved Plasmodium protein, unknown function
Length=1069
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 30/55 (54%), Gaps = 6/55 (10%)
Query 3 HVYVQLLNTAYMRAYVFK-----NFLFEVPALNLHLFAKDLRKIIPSIQVEDLTY 52
++Y +LNT Y + Y F NF + + +N++ KDL ++ + +E+L Y
Sbjct 365 NLYCSILNTVYKKIYFFPCIDIINFWYIIKRINIYSI-KDLSSLVINKLMENLKY 418
> pfa:PF13_0155 conserved Plasmodium protein, unknown function
Length=2668
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 23/83 (27%), Positives = 34/83 (40%), Gaps = 17/83 (20%)
Query 1 LAHVYVQLLN------TAYMRAYVFKNFLFEV-----------PALNLHLFAKDLRKIIP 43
L +V+VQ N ++ +V +FLF ALN H+F K I
Sbjct 2212 LFNVFVQKSNHNNNKFNQFLDDHVLDDFLFNTFNGKIHGDNIKRALNYHMFESRFLKDIT 2271
Query 44 SIQVEDLTYAKGFGGIRPQLINK 66
I +D+TY K I ++ K
Sbjct 2272 KINNDDITYDKYLQCIHASVLKK 2294
> ath:AT2G13540 ABH1; ABH1 (ABA HYPERSENSITIVE 1); RNA cap binding;
K12882 nuclear cap-binding protein subunit 1
Length=848
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query 37 DLRKIIPSIQVEDLTYAKGFGGIRPQLINKTEKKLLLGEGKIVTGEN 83
DLRK I +I L K R +L E KL L EG+ V GEN
Sbjct 646 DLRKDISNITKNVLVAEKASANARVEL-EAAESKLSLVEGEPVLGEN 691
Lambda K H
0.323 0.142 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012750684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40