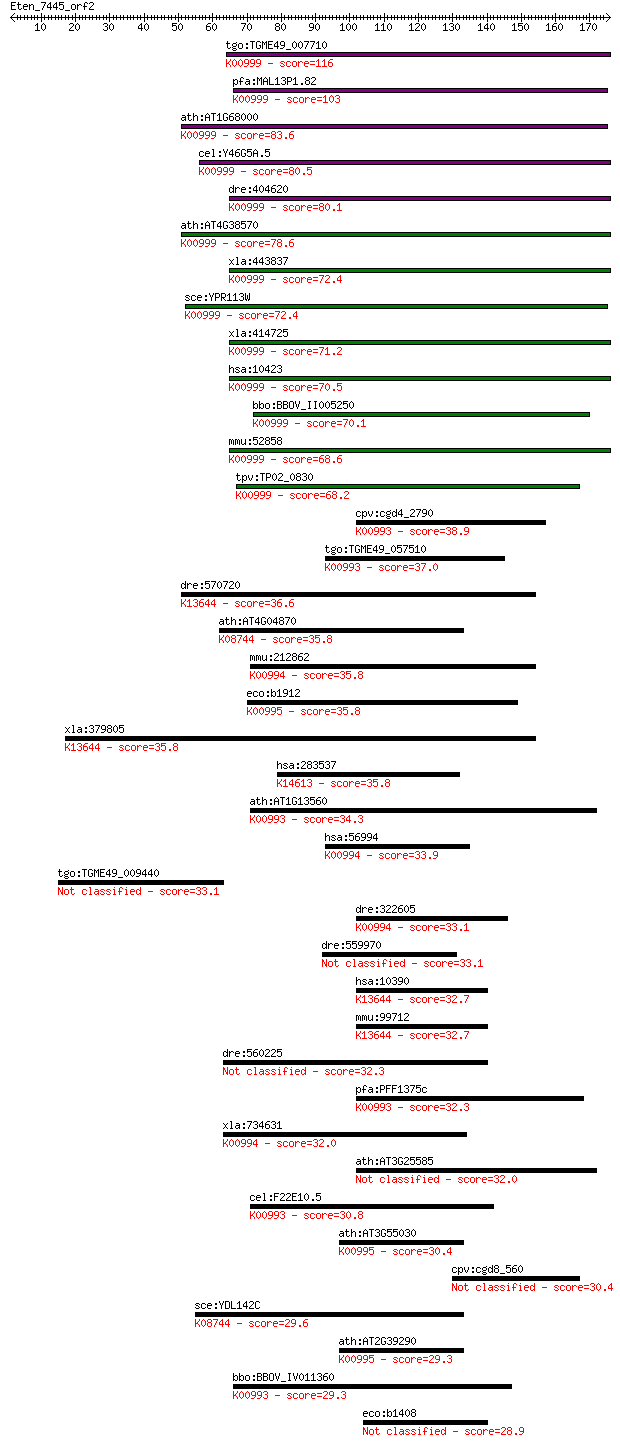
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7445_orf2
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_007710 phosphatidylinositol synthase, putative (EC:... 116 4e-26
pfa:MAL13P1.82 phosphatidylinositol synthase (EC:2.7.8.11); K0... 103 3e-22
ath:AT1G68000 ATPIS1; ATPIS1 (PHOSPHATIDYLINOSITOL SYNTHASE 1)... 83.6 3e-16
cel:Y46G5A.5 pisy-1; PhosphatidylInositol SYnthase family memb... 80.5 3e-15
dre:404620 cdipt, MGC174794, MGC77540, zgc:77540; CDP-diacylgl... 80.1 3e-15
ath:AT4G38570 PIS2; PIS2 (PROBABLE CDP-DIACYLGLYCEROL--INOSITO... 78.6 9e-15
xla:443837 cdipt, MGC82599; CDP-diacylglycerol--inositol 3-pho... 72.4 7e-13
sce:YPR113W PIS1; Phosphatidylinositol synthase, required for ... 72.4 7e-13
xla:414725 hypothetical protein MGC81124; K00999 CDP-diacylgly... 71.2 2e-12
hsa:10423 CDIPT, MGC1328, PIS, PIS1; CDP-diacylglycerol--inosi... 70.5 3e-12
bbo:BBOV_II005250 18.m06434; phosphatidylinositol synthase (EC... 70.1 4e-12
mmu:52858 Cdipt, 9530042F15Rik, D7Bwg0575e, Pis, Pis1; CDP-dia... 68.6 1e-11
tpv:TP02_0830 phosphatidylinositol synthase; K00999 CDP-diacyl... 68.2 1e-11
cpv:cgd4_2790 ethanolaminephosphotransferase (ETHPT) 9 transme... 38.9 0.010
tgo:TGME49_057510 aminoalcoholphosphotransferase, putative (EC... 37.0 0.034
dre:570720 cept1, zgc:171762; choline/ethanolamine phosphotran... 36.6 0.045
ath:AT4G04870 CLS; CLS (CARDIOLIPIN SYNTHASE); cardiolipin syn... 35.8 0.069
mmu:212862 Chpt1, MGC28885; choline phosphotransferase 1 (EC:2... 35.8 0.073
eco:b1912 pgsA, ECK1911, JW1897; phosphatidylglycerophosphate ... 35.8 0.074
xla:379805 cept1, MGC53412; choline/ethanolamine phosphotransf... 35.8 0.079
hsa:283537 SLC46A3, FKSG16, FLJ42613; solute carrier family 46... 35.8 0.083
ath:AT1G13560 AAPT1; AAPT1 (AMINOALCOHOLPHOSPHOTRANSFERASE 1);... 34.3 0.21
hsa:56994 CHPT1, CPT, CPT1; choline phosphotransferase 1 (EC:2... 33.9 0.32
tgo:TGME49_009440 hypothetical protein 33.1 0.48
dre:322605 chpt1, fd50f07, sycp3, wu:fb67a04, wu:fd50f07; chol... 33.1 0.55
dre:559970 si:dkey-205h13.1 33.1 0.56
hsa:10390 CEPT1, DKFZp313G0615, MGC45223; choline/ethanolamine... 32.7 0.57
mmu:99712 Cept1, 9930118K05Rik, AI788721, BB118941; choline/et... 32.7 0.58
dre:560225 si:dkey-208b23.1 32.3 0.80
pfa:PFF1375c ethanolaminephosphotransferase, putative (EC:2.7.... 32.3 0.86
xla:734631 chpt1, MGC114982; choline phosphotransferase 1 (EC:... 32.0 1.0
ath:AT3G25585 AAPT2; AAPT2 (AMINOALCOHOLPHOSPHOTRANSFERASE); p... 32.0 1.1
cel:F22E10.5 hypothetical protein; K00993 ethanolaminephosphot... 30.8 2.3
ath:AT3G55030 PGPS2; PGPS2 (phosphatidylglycerolphosphate synt... 30.4 2.9
cpv:cgd8_560 CDP-diacylglycerol--inositol 3-phosphatidyltransf... 30.4 3.2
sce:YDL142C CRD1, CLS1; Cardiolipin synthase; produces cardiol... 29.6 4.8
ath:AT2G39290 PGP1; PGP1 (PHOSPHATIDYLGLYCEROLPHOSPHATE SYNTHA... 29.3 6.2
bbo:BBOV_IV011360 23.m05792; ethanolamine phosphatidyltransfer... 29.3 6.7
eco:b1408 ynbA, ECK1401, JW1405; inner membrane protein 28.9 8.1
> tgo:TGME49_007710 phosphatidylinositol synthase, putative (EC:2.7.8.11);
K00999 CDP-diacylglycerol--inositol 3-phosphatidyltransferase
[EC:2.7.8.11]
Length=258
Score = 116 bits (290), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 68/115 (59%), Positives = 83/115 (72%), Gaps = 3/115 (2%)
Query 64 LKVLLYVPNLIGYLRLLLLLVAALVGSQ---WLCLLCYAASQLLDAADGHTARRLNQVSL 120
LKV LYVPN+IGY+R+ LLL AA + L + Y SQ LDA DG ARRL QVS+
Sbjct 47 LKVFLYVPNIIGYVRIALLLAAAFAWREAYTSLFFVSYVTSQCLDAVDGAAARRLGQVSI 106
Query 121 FGACLDQLLDRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLHFFAATAAGA 175
GACLDQ++DRLSTCL Y+LNA+V P +A+ FF+ L D+GGHW+HFFAA GA
Sbjct 107 VGACLDQVVDRLSTCLLYVLNASVYPDFAFAFFIALFLDMGGHWVHFFAAAVVGA 161
> pfa:MAL13P1.82 phosphatidylinositol synthase (EC:2.7.8.11);
K00999 CDP-diacylglycerol--inositol 3-phosphatidyltransferase
[EC:2.7.8.11]
Length=209
Score = 103 bits (257), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 51/111 (45%), Positives = 73/111 (65%), Gaps = 2/111 (1%)
Query 66 VLLYVPNLIGYLRLLLLLVAALVGSQWLCLLC--YAASQLLDAADGHTARRLNQVSLFGA 123
V LY+PN+IGY+R++L L+ ++ + L L Y+ SQ+LDA DG TAR+ NQ S+FG
Sbjct 8 VYLYIPNIIGYIRVILALLGFIISRKNLFLFVCFYSVSQVLDALDGWTARKFNQTSVFGQ 67
Query 124 CLDQLLDRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLHFFAATAAG 174
LDQ+ DRLST L YLLN++V Y L+++ D+ GH+ H + AG
Sbjct 68 ILDQITDRLSTSLLYLLNSSVYEEYITLIGLIMIADIAGHYFHSTSCAIAG 118
> ath:AT1G68000 ATPIS1; ATPIS1 (PHOSPHATIDYLINOSITOL SYNTHASE
1); CDP-diacylglycerol-inositol 3-phosphatidyltransferase; K00999
CDP-diacylglycerol--inositol 3-phosphatidyltransferase
[EC:2.7.8.11]
Length=227
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 55/127 (43%), Positives = 72/127 (56%), Gaps = 7/127 (5%)
Query 51 MAQQTPPRGDTEVLKVLLYVPNLIGYLRLLLLLVAALV--GSQWLCLLCYAASQLLDAAD 108
MA++ PR E L V LY+PN++GY+R+LL VA V ++ L + Y S DA D
Sbjct 1 MAKKERPRP--EKLSVYLYIPNIVGYMRVLLNCVAFAVCFSNKPLFSVLYFFSFCCDAVD 58
Query 109 GHTARRLNQVSLFGACLDQLLDRLST-CLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLHF 167
G ARR NQVS FGA LD + DR+ST CL +L+ P F LL D+ HWL
Sbjct 59 GWVARRFNQVSTFGAVLDMVTDRVSTACLLVILSQIYRPSLV--FLSLLALDIASHWLQM 116
Query 168 FAATAAG 174
++ AG
Sbjct 117 YSTFLAG 123
> cel:Y46G5A.5 pisy-1; PhosphatidylInositol SYnthase family member
(pisy-1); K00999 CDP-diacylglycerol--inositol 3-phosphatidyltransferase
[EC:2.7.8.11]
Length=220
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 54/125 (43%), Positives = 64/125 (51%), Gaps = 8/125 (6%)
Query 56 PPRGDTEVLKVLLYVPNLIGYLRLLLLLVAALVGSQ--WLCLLCYAASQLLDAADGHTAR 113
P V L+ PNLIGY R++L ++A S W + CYA S LDA DG AR
Sbjct 2 EPTAAENNNNVWLFYPNLIGYGRIVLAILAMYYMSSAPWCAMFCYALSAGLDAFDGWAAR 61
Query 114 RLNQVSLFGACLDQLLDRLSTCLFYLLNAAVCPHYAWGFFLLLLT---DVGGHWLHFFAA 170
NQ S FGA LDQL DR C L A+C Y FLL L+ D+ HWLH A
Sbjct 62 TYNQSSRFGAMLDQLTDR---CALLALVMALCKFYPDHLFLLQLSAVIDIASHWLHLHAT 118
Query 171 TAAGA 175
+GA
Sbjct 119 DLSGA 123
> dre:404620 cdipt, MGC174794, MGC77540, zgc:77540; CDP-diacylglycerol--inositol
3-phosphatidyltransferase (phosphatidylinositol
synthase) (EC:2.7.8.11); K00999 CDP-diacylglycerol--inositol
3-phosphatidyltransferase [EC:2.7.8.11]
Length=213
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 50/116 (43%), Positives = 65/116 (56%), Gaps = 8/116 (6%)
Query 65 KVLLYVPNLIGYLRLLLLLVAALVGSQWLC-----LLCYAASQLLDAADGHTARRLNQVS 119
+ L+VPNLIGY R++L LV+ + C + CY S LLDA DGH AR LNQ +
Sbjct 5 NIFLFVPNLIGYARIVLALVSFFLMP---CCPGPAVFCYLLSALLDAFDGHAARALNQGT 61
Query 120 LFGACLDQLLDRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLHFFAATAAGA 175
FGA LD L DR +T + A + P Y + F L + DV HWLH ++ GA
Sbjct 62 KFGAMLDMLTDRCATMCLLVNLALLYPSYTFLFQLSMCLDVASHWLHLHSSMMKGA 117
> ath:AT4G38570 PIS2; PIS2 (PROBABLE CDP-DIACYLGLYCEROL--INOSITOL
3-PHOSPHATIDYLTRANSFERASE 2); phosphotransferase, for other
substituted phosphate groups; K00999 CDP-diacylglycerol--inositol
3-phosphatidyltransferase [EC:2.7.8.11]
Length=225
Score = 78.6 bits (192), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 70/128 (54%), Gaps = 10/128 (7%)
Query 51 MAQQTPPRGDTEVLKVLLYVPNLIGYLRLLLLLVAALV--GSQWLCLLCYAASQLLDAAD 108
MA+Q P L V LY+PN++GY+R+LL +A V ++ L L Y S DA D
Sbjct 1 MAKQRPA-----TLSVYLYIPNIVGYMRVLLNCIAFSVCFSNKTLFSLLYFFSFCCDAVD 55
Query 109 GHTARRLNQVSLFGACLDQLLDRLST-CLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLHF 167
G AR+ NQVS FGA LD + DR+ST CL +L+ P F LL D+ HWL
Sbjct 56 GWCARKFNQVSTFGAVLDMVTDRVSTACLLVILSQIYRPSLV--FLSLLALDIASHWLQM 113
Query 168 FAATAAGA 175
++ +G
Sbjct 114 YSTFLSGK 121
> xla:443837 cdipt, MGC82599; CDP-diacylglycerol--inositol 3-phosphatidyltransferase
(EC:2.7.8.11); K00999 CDP-diacylglycerol--inositol
3-phosphatidyltransferase [EC:2.7.8.11]
Length=218
Score = 72.4 bits (176), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 44/113 (38%), Positives = 61/113 (53%), Gaps = 2/113 (1%)
Query 65 KVLLYVPNLIGYLRLLLLLVAA--LVGSQWLCLLCYAASQLLDAADGHTARRLNQVSLFG 122
+ L+VPNLIGY R++ +A + S + Y S LLDA DGH AR LNQ + FG
Sbjct 5 NIFLFVPNLIGYARIVFAFIAFYFMPTSPVMASTFYLVSGLLDAFDGHAARLLNQGTKFG 64
Query 123 ACLDQLLDRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLHFFAATAAGA 175
A LD L DR +T + + + P Y F L + D+ HWLH ++ G+
Sbjct 65 AMLDMLTDRCATMCLLVNLSLLYPSYTLLFQLSMSLDIASHWLHLHSSILKGS 117
> sce:YPR113W PIS1; Phosphatidylinositol synthase, required for
biosynthesis of phosphatidylinositol, which is a precursor
for polyphosphoinositides, sphingolipids, and glycolipid anchors
for some of the plasma membrane proteins (EC:2.7.8.11);
K00999 CDP-diacylglycerol--inositol 3-phosphatidyltransferase
[EC:2.7.8.11]
Length=220
Score = 72.4 bits (176), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 47/129 (36%), Positives = 66/129 (51%), Gaps = 12/129 (9%)
Query 52 AQQTPPRGDTEVLKVLLYVPNLIGYLRLLLLLVAALVGSQ------WLCLLCYAASQLLD 105
+ TP + E VL Y+PN IGY+R++ ++ V WL Y+ S LLD
Sbjct 3 SNSTPEKVTAE--HVLWYIPNKIGYVRVITAALSFFVMKNHPTAFTWL----YSTSCLLD 56
Query 106 AADGHTARRLNQVSLFGACLDQLLDRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWL 165
A DG AR+ NQVS GA LD + DR ST P + F L+L D+ H++
Sbjct 57 ALDGTMARKYNQVSSLGAVLDMVTDRSSTAGLMCFLCVQYPQWCVFFQLMLGLDITSHYM 116
Query 166 HFFAATAAG 174
H +A+ +AG
Sbjct 117 HMYASLSAG 125
> xla:414725 hypothetical protein MGC81124; K00999 CDP-diacylglycerol--inositol
3-phosphatidyltransferase [EC:2.7.8.11]
Length=218
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 45/113 (39%), Positives = 61/113 (53%), Gaps = 2/113 (1%)
Query 65 KVLLYVPNLIGYLRLLLLLVAA--LVGSQWLCLLCYAASQLLDAADGHTARRLNQVSLFG 122
V L+VPNLIGY R++ +A + S + Y S LLDA DGH AR LNQ + FG
Sbjct 5 NVFLFVPNLIGYARIVFAFLAFYFMPTSPIMASTFYLLSGLLDAFDGHAARLLNQGTKFG 64
Query 123 ACLDQLLDRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLHFFAATAAGA 175
A LD L DR +T + + + P Y F L + D+ HWLH ++ G+
Sbjct 65 AMLDMLTDRCATMCLLVNLSLLYPSYTLLFQLSMSLDIASHWLHLHSSILKGS 117
> hsa:10423 CDIPT, MGC1328, PIS, PIS1; CDP-diacylglycerol--inositol
3-phosphatidyltransferase (EC:2.7.8.11); K00999 CDP-diacylglycerol--inositol
3-phosphatidyltransferase [EC:2.7.8.11]
Length=213
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 50/120 (41%), Positives = 67/120 (55%), Gaps = 16/120 (13%)
Query 65 KVLLYVPNLIGYLRLLLLLVA-------ALVGSQWLCLLCYAASQLLDAADGHTARRLNQ 117
+ L+VPNLIGY R++ +++ L S + Y S LLDA DGH AR LNQ
Sbjct 5 NIFLFVPNLIGYARIVFAIISFYFMPCCPLTASSF-----YLLSGLLDAFDGHAARALNQ 59
Query 118 VSLFGACLDQLLDRLST-CLFYLLNAAVCPHYAWGFFLLLLT-DVGGHWLHFFAATAAGA 175
+ FGA LD L DR ST CL L+N A+ A FF + ++ DV HWLH ++ G+
Sbjct 60 GTRFGAMLDMLTDRCSTMCL--LVNLALLYPGATLFFQISMSLDVASHWLHLHSSVVRGS 117
> bbo:BBOV_II005250 18.m06434; phosphatidylinositol synthase (EC:2.7.8.11);
K00999 CDP-diacylglycerol--inositol 3-phosphatidyltransferase
[EC:2.7.8.11]
Length=208
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 36/100 (36%), Positives = 58/100 (58%), Gaps = 2/100 (2%)
Query 72 NLIGYLRLLLLLVAALVGSQ--WLCLLCYAASQLLDAADGHTARRLNQVSLFGACLDQLL 129
N + +R +L+++ + ++ + Y AS LLD DG+ +R N+ +L GA DQLL
Sbjct 12 NGVTMIRTILIMIGMFFARRNPFVFVSLYTASHLLDMLDGYVSRYYNESTLVGAAFDQLL 71
Query 130 DRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLHFFA 169
DR+S+ LNA P F+L++L D+ GHW+H +A
Sbjct 72 DRMSSTYLCFLNARQYPRCLEVFYLIMLIDICGHWIHNYA 111
> mmu:52858 Cdipt, 9530042F15Rik, D7Bwg0575e, Pis, Pis1; CDP-diacylglycerol--inositol
3-phosphatidyltransferase (phosphatidylinositol
synthase) (EC:2.7.8.11); K00999 CDP-diacylglycerol--inositol
3-phosphatidyltransferase [EC:2.7.8.11]
Length=213
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 44/118 (37%), Positives = 60/118 (50%), Gaps = 12/118 (10%)
Query 65 KVLLYVPNLIGYLRLLLLLVA-------ALVGSQWLCLLCYAASQLLDAADGHTARRLNQ 117
+ L+VPNLIGY R++ +++ S + Y S LLDA DGH AR LNQ
Sbjct 5 NIFLFVPNLIGYARIVFAIISFYFMPCCPFTASSF-----YLLSGLLDAFDGHAARALNQ 59
Query 118 VSLFGACLDQLLDRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLHFFAATAAGA 175
+ FGA LD L DR +T + A + P F L + DV HWLH ++ G+
Sbjct 60 GTRFGAMLDMLTDRCATMCLLVNLALLYPRATLLFQLSMSLDVASHWLHLHSSVVRGS 117
> tpv:TP02_0830 phosphatidylinositol synthase; K00999 CDP-diacylglycerol--inositol
3-phosphatidyltransferase [EC:2.7.8.11]
Length=214
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 39/102 (38%), Positives = 55/102 (53%), Gaps = 2/102 (1%)
Query 67 LLYVPNLIGYLRLLLLLVAALV-GSQWLCLL-CYAASQLLDAADGHTARRLNQVSLFGAC 124
LL PN I R+LL + L ++ L L Y +S +LD DG AR+ + ++FGA
Sbjct 8 LLNKPNFITSFRVLLFCTSFLYYKTKPLYFLGFYVSSVVLDMVDGEVARKYGESTIFGAM 67
Query 125 LDQLLDRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLH 166
D L DR++T Y+ A P Y F+ +L+ DV GHW H
Sbjct 68 FDSLFDRMTTAFVYMHLAMKYPKYFLFFYFVLVWDVVGHWAH 109
> cpv:cgd4_2790 ethanolaminephosphotransferase (ETHPT) 9 transmembrane
domain protein involved in lipid ; K00993 ethanolaminephosphotransferase
[EC:2.7.8.1]
Length=428
Score = 38.9 bits (89), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 102 QLLDAADGHTARRLNQVSLFGACLDQLLDRLSTCLFYLLNAAVCPH-YAWGFFLLL 156
Q DAADG ARRL S G LD LD +T F + A C +++ FF+ L
Sbjct 106 QTFDAADGKHARRLKISSPLGQLLDHGLDSYTTIFFSTIFCACCKMGWSYKFFIFL 161
> tgo:TGME49_057510 aminoalcoholphosphotransferase, putative (EC:2.7.8.1);
K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=467
Score = 37.0 bits (84), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 25/52 (48%), Gaps = 1/52 (1%)
Query 93 LCLLCYAASQLLDAADGHTARRLNQVSLFGACLDQLLDRLSTCLFYLLNAAV 144
+CLL + Q LDA DG ARR N + G D D ST +NAA
Sbjct 90 MCLLLFV-YQTLDAVDGKQARRTNSSTPLGQLFDHGCDSFSTVFAVFINAAT 140
> dre:570720 cept1, zgc:171762; choline/ethanolamine phosphotransferase
1; K13644 choline/ethanolamine phosphotransferase
[EC:2.7.8.1 2.7.8.2]
Length=415
Score = 36.6 bits (83), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 34/113 (30%), Positives = 48/113 (42%), Gaps = 13/113 (11%)
Query 51 MAQQTPPRGDTEVLKVLLYVPNLIGYLRLLLLLVAALVGSQ---WLCLLCYA---ASQLL 104
+ + PP ++ ++ V N+ + L+L+ Q W LLC Q L
Sbjct 75 LVNRMPPWIAPNLITIVGLVTNI--FTTLVLVYYCPTATEQAPLWAYLLCAVGLFVYQSL 132
Query 105 DAADGHTARRLNQVSLFGACLDQLLDRLSTCLFYLLNAAVC----PHYAWGFF 153
DA DG ARR N S G D D LST +F +L ++ H W FF
Sbjct 133 DAIDGKQARRTNSSSPLGELFDHGCDSLST-VFVVLGTSIAVQLGTHPDWMFF 184
> ath:AT4G04870 CLS; CLS (CARDIOLIPIN SYNTHASE); cardiolipin synthase/
phosphatidyltransferase; K08744 cardiolipin synthase
[EC:2.7.8.-]
Length=341
Score = 35.8 bits (81), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 40/77 (51%), Gaps = 9/77 (11%)
Query 62 EVLKVLLYVPNLIGYLRLL------LLLVAALVGSQWLCLLCYAASQLLDAADGHTARRL 115
+++K + VPN+I RL+ ++ + S +L L AS D DG+ ARR+
Sbjct 136 KLVKSFVNVPNMISMARLVSGPVLWWMISNEMYSSAFLGLAVSGAS---DWLDGYVARRM 192
Query 116 NQVSLFGACLDQLLDRL 132
S+ G+ LD L D++
Sbjct 193 KINSVVGSYLDPLADKV 209
> mmu:212862 Chpt1, MGC28885; choline phosphotransferase 1 (EC:2.7.8.2);
K00994 diacylglycerol cholinephosphotransferase [EC:2.7.8.2]
Length=406
Score = 35.8 bits (81), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 41/101 (40%), Gaps = 19/101 (18%)
Query 71 PNLIGYLRLLLLLVAALV-----------GSQWLCLLCYAA---SQLLDAADGHTARRLN 116
PN I + L + LV LV W LLC Q LDA DG ARR N
Sbjct 63 PNTITLIGLAINLVTTLVLIFYCPTVTEEAPYWTYLLCALGLFIYQSLDAIDGKQARRTN 122
Query 117 QVSLFGACLDQLLDRLSTCLFYLLNAAVC----PHYAWGFF 153
S G D D LST +F + A++ H W FF
Sbjct 123 SCSPLGELFDHGCDSLST-VFMAIGASIAVRLGTHPDWLFF 162
> eco:b1912 pgsA, ECK1911, JW1897; phosphatidylglycerophosphate
synthetase (EC:2.7.8.5); K00995 CDP-diacylglycerol--glycerol-3-phosphate
3-phosphatidyltransferase [EC:2.7.8.5]
Length=182
Score = 35.8 bits (81), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 41/85 (48%), Gaps = 9/85 (10%)
Query 70 VPNLIGYLRLLLLLVAALV------GSQWLCLLCYAASQLLDAADGHTARRLNQVSLFGA 123
+P L+ R++L+ LV S + L + + + D DG ARR NQ + FGA
Sbjct 5 IPTLLTLFRVILIPFFVLVFYLPVTWSPFAAALIFCVAAVTDWFDGFLARRWNQSTRFGA 64
Query 124 CLDQLLDRLSTCLFYLLNAAVCPHY 148
LD + D++ + +L V HY
Sbjct 65 FLDPVADKVLVAIAMVL---VTEHY 86
> xla:379805 cept1, MGC53412; choline/ethanolamine phosphotransferase
1 (EC:2.7.8.2 2.7.8.1); K13644 choline/ethanolamine
phosphotransferase [EC:2.7.8.1 2.7.8.2]
Length=416
Score = 35.8 bits (81), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 49/166 (29%), Positives = 64/166 (38%), Gaps = 32/166 (19%)
Query 17 CGNPSAAA-------RSPAPPLRGPQLPAAALSSSAFYLFTMAQQTP-PRGDTE--VLKV 66
CGNP A P PPL QL L + + P +G E V++V
Sbjct 22 CGNPYQTACLLSKFIELPNPPLSRHQL--KRLEEHRYQSCGKSLLEPLMQGFWEWLVIQV 79
Query 67 LLYV-PNLIGYLRLLLLLVAALV-----------GSQWLCLLCYAA---SQLLDAADGHT 111
++ PNLI + LL+ +V +V W L C Q LDA DG
Sbjct 80 PQWIAPNLITIIGLLINIVTTVVLIYYCPTATEKAPTWTYLSCAIGLFMYQSLDAIDGKQ 139
Query 112 ARRLNQVSLFGACLDQLLDRLSTCLFYLLNAAVCPHYA----WGFF 153
ARR N + G D D LST +F +L + W FF
Sbjct 140 ARRTNSSTPLGELFDHGCDSLST-VFVVLGTCIAVQLGTNPDWMFF 184
> hsa:283537 SLC46A3, FKSG16, FLJ42613; solute carrier family
46, member 3; K14613 MFS transporter, PCFT/HCP family, solute
carrier family 46 (folate transporter), member 1/3
Length=463
Score = 35.8 bits (81), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 31/55 (56%), Gaps = 2/55 (3%)
Query 79 LLLLLVAALVGSQWLCLLCYAA--SQLLDAADGHTARRLNQVSLFGACLDQLLDR 131
++L V AL S WLCLLCY A QLL A+ A N + +GAC ++D+
Sbjct 104 MILSSVGALATSVWLCLLCYFAFPFQLLIASTFIGAFCGNYTTFWGACFAYIVDQ 158
> ath:AT1G13560 AAPT1; AAPT1 (AMINOALCOHOLPHOSPHOTRANSFERASE 1);
phosphatidyltransferase/ phosphotransferase, for other substituted
phosphate groups (EC:2.7.8.1); K00993 ethanolaminephosphotransferase
[EC:2.7.8.1]
Length=389
Score = 34.3 bits (77), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 52/122 (42%), Gaps = 25/122 (20%)
Query 71 PNLIGYLRLLLLLVAALVG-----------SQW------LCLLCYAASQLLDAADGHTAR 113
PN+I + + L+ ++L+G +W L L Y Q DA DG AR
Sbjct 47 PNMITLMGFMFLVTSSLLGYIYSPQLDSPPPRWVHFAHGLLLFLY---QTFDAVDGKQAR 103
Query 114 RLNQVSLFGACLDQLLDRLSTCLFYLL---NAAVCPHYAWGFFLL-LLTDVGGHWLHFFA 169
R N S G D D L+ C F + + A+C + F+++ + G W H+F
Sbjct 104 RTNSSSPLGELFDHGCDALA-CAFEAMAFGSTAMCGRDTFWFWVISAIPFYGATWEHYFT 162
Query 170 AT 171
T
Sbjct 163 NT 164
> hsa:56994 CHPT1, CPT, CPT1; choline phosphotransferase 1 (EC:2.7.8.2);
K00994 diacylglycerol cholinephosphotransferase [EC:2.7.8.2]
Length=406
Score = 33.9 bits (76), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 93 LCLLCYAASQLLDAADGHTARRLNQVSLFGACLDQLLDRLST 134
LC L Q LDA DG ARR N S G D D LST
Sbjct 99 LCALGLFIYQSLDAIDGKQARRTNSCSPLGELFDHGCDSLST 140
> tgo:TGME49_009440 hypothetical protein
Length=3407
Score = 33.1 bits (74), Expect = 0.48, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 26/58 (44%), Gaps = 10/58 (17%)
Query 15 HCCGNPSAAARSPAPPLRGPQLP--------AAALSSSAFYLFTM--AQQTPPRGDTE 62
H G P +AR PPL GP+ P L S A F + AQ P RG++E
Sbjct 2896 HAGGAPLTSARPLGPPLAGPKAPREEPREASVGLLGSEATPAFKLVQAQVDPHRGESE 2953
> dre:322605 chpt1, fd50f07, sycp3, wu:fb67a04, wu:fd50f07; choline
phosphotransferase 1 (EC:2.7.8.2); K00994 diacylglycerol
cholinephosphotransferase [EC:2.7.8.2]
Length=382
Score = 33.1 bits (74), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 22/44 (50%), Gaps = 1/44 (2%)
Query 102 QLLDAADGHTARRLNQVSLFGACLDQLLDRLSTCLFYLLNAAVC 145
Q LDA DG ARR N S G D D +ST +F + +C
Sbjct 97 QSLDAIDGKQARRTNSSSALGELFDHGCDAVST-VFVAVGTCIC 139
> dre:559970 si:dkey-205h13.1
Length=241
Score = 33.1 bits (74), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 21/39 (53%), Gaps = 4/39 (10%)
Query 92 WLCLLCYAASQLLDAADGHTARRLNQVSLFGACLDQLLD 130
WL L+ Y LLD ADG AR LN S GA LD D
Sbjct 65 WLVLIGY----LLDLADGAVARHLNACSALGAKLDDFAD 99
> hsa:10390 CEPT1, DKFZp313G0615, MGC45223; choline/ethanolamine
phosphotransferase 1 (EC:2.7.8.2 2.7.8.1); K13644 choline/ethanolamine
phosphotransferase [EC:2.7.8.1 2.7.8.2]
Length=416
Score = 32.7 bits (73), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 18/38 (47%), Gaps = 0/38 (0%)
Query 102 QLLDAADGHTARRLNQVSLFGACLDQLLDRLSTCLFYL 139
Q LDA DG ARR N S G D D LST L
Sbjct 130 QSLDAIDGKQARRTNSSSPLGELFDHGCDSLSTVFVVL 167
> mmu:99712 Cept1, 9930118K05Rik, AI788721, BB118941; choline/ethanolaminephosphotransferase
1 (EC:2.7.8.2 2.7.8.1); K13644
choline/ethanolamine phosphotransferase [EC:2.7.8.1 2.7.8.2]
Length=416
Score = 32.7 bits (73), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 18/38 (47%), Gaps = 0/38 (0%)
Query 102 QLLDAADGHTARRLNQVSLFGACLDQLLDRLSTCLFYL 139
Q LDA DG ARR N S G D D LST L
Sbjct 130 QSLDAIDGKQARRTNSSSPLGELFDHGCDSLSTVFVVL 167
> dre:560225 si:dkey-208b23.1
Length=426
Score = 32.3 bits (72), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 38/92 (41%), Gaps = 15/92 (16%)
Query 63 VLKVLLYV-PNLIGYLRLLLLLVAALV-----------GSQWLCLLCYAA---SQLLDAA 107
V KV L++ PNLI + L +V+ L+ W L C Q LDA
Sbjct 80 VSKVPLWMAPNLITIVGLATNIVSTLILVYYCPTATEQAPTWAYLACALGLFIYQSLDAI 139
Query 108 DGHTARRLNQVSLFGACLDQLLDRLSTCLFYL 139
DG ARR N + G D D LST L
Sbjct 140 DGKQARRTNSSTPLGELFDHGCDSLSTVFVVL 171
> pfa:PFF1375c ethanolaminephosphotransferase, putative (EC:2.7.8.1);
K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=391
Score = 32.3 bits (72), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 31/71 (43%), Gaps = 5/71 (7%)
Query 102 QLLDAADGHTARRLNQVSLFGACLDQLLDRLSTCLFYLL--NAAVCPHYAWGFFLLLLTD 159
Q DA DG AR+ N S G D D ++T F + AA P + LL +
Sbjct 92 QTFDALDGKQARKTNTSSPLGQLFDHGCDSITTSFFVFIACKAAGFPKSLIYYILLAIVQ 151
Query 160 VGGH---WLHF 167
+ G+ W+ +
Sbjct 152 IQGYMFSWMEY 162
> xla:734631 chpt1, MGC114982; choline phosphotransferase 1 (EC:2.7.8.2);
K00994 diacylglycerol cholinephosphotransferase
[EC:2.7.8.2]
Length=402
Score = 32.0 bits (71), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 38/86 (44%), Gaps = 15/86 (17%)
Query 63 VLKVLLYV-PNLIGYLRLLLLLVAALV-----------GSQWLCLLCYA---ASQLLDAA 107
V KV L++ PN I + LLL +++ L+ W LLC Q LDA
Sbjct 54 VEKVPLWLAPNTITMVGLLLNVLSTLILVCYCPTATEGAPFWTYLLCAIGLFVYQSLDAI 113
Query 108 DGHTARRLNQVSLFGACLDQLLDRLS 133
DG ARR N S G D D +S
Sbjct 114 DGKQARRTNSSSPLGEMFDHGCDSIS 139
> ath:AT3G25585 AAPT2; AAPT2 (AMINOALCOHOLPHOSPHOTRANSFERASE);
phosphatidyltransferase/ phosphotransferase, for other substituted
phosphate groups
Length=337
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 32/73 (43%), Gaps = 3/73 (4%)
Query 102 QLLDAADGHTARRLNQVSLFGACLDQLLDRLSTCLFYLL--NAAVCPHYAWGFFLL-LLT 158
Q DA DG ARR N S G D D L L + + A+C + F+++ +
Sbjct 40 QTFDAVDGKQARRTNSSSPLGELFDHGCDALGCALETMAYGSTAMCGRDTFWFWVISAVP 99
Query 159 DVGGHWLHFFAAT 171
G W H+F T
Sbjct 100 FFGATWEHYFTNT 112
> cel:F22E10.5 hypothetical protein; K00993 ethanolaminephosphotransferase
[EC:2.7.8.1]
Length=394
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 28/89 (31%), Positives = 37/89 (41%), Gaps = 23/89 (25%)
Query 71 PNLIGYLRLLLLLVAALVGSQWLCLLCYAAS------------------QLLDAADGHTA 112
PNLI ++ L++ L+ LV S Y+A+ Q LDA DG A
Sbjct 66 PNLITWIGLVINLITVLVLSS----FSYSATESAPSWAYLQAALGLFFYQTLDAIDGKQA 121
Query 113 RRLNQVSLFGACLDQLLDRLSTCLFYLLN 141
RR S G D D + T +F LN
Sbjct 122 RRTGSSSPLGELFDHGCDSM-TQVFVTLN 149
> ath:AT3G55030 PGPS2; PGPS2 (phosphatidylglycerolphosphate synthase
2); CDP-alcohol phosphatidyltransferase/ CDP-diacylglycerol-glycerol-3-phosphate
3-phosphatidyltransferase (EC:2.7.8.5);
K00995 CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase
[EC:2.7.8.5]
Length=233
Score = 30.4 bits (67), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 97 CYAASQLLDAADGHTARRLNQVSLFGACLDQLLDRL 132
+ A+ + D DG+ AR++ S FGA LD + D+L
Sbjct 76 IFIAAAITDWLDGYIARKMRLGSEFGAFLDPVADKL 111
> cpv:cgd8_560 CDP-diacylglycerol--inositol 3-phosphatidyltransferase
isoform 1; phosphatidylinositol synthase; PtdIns synthase;
PI synthase
Length=154
Score = 30.4 bits (67), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 130 DRLSTCLFYLLNAAVCPHYAWGFFLLLLTDVGGHWLH 166
DR ST + +L + Y + + L+ D+ GHWL+
Sbjct 4 DRCSTVIIIILAITLNKSYTFLMIIFLIGDISGHWLY 40
> sce:YDL142C CRD1, CLS1; Cardiolipin synthase; produces cardiolipin,
which is a phospholipid of the mitochondrial inner membrane
that is required for normal mitochondrial membrane potential
and function; also required for normal vacuolar ion
homeostasis (EC:2.7.8.-); K08744 cardiolipin synthase [EC:2.7.8.-]
Length=283
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 38/85 (44%), Gaps = 11/85 (12%)
Query 55 TPPRGDTEVLKVLLYVPNLIGYLRL-------LLLLVAALVGSQWLCLLCYAASQLLDAA 107
T P + LL +PN++ R+ L ++ L + L +A S + D
Sbjct 55 TNPSKTPHIKSKLLNIPNILTLSRIGCTPFIGLFIITNNLTPA----LGLFAFSSITDFM 110
Query 108 DGHTARRLNQVSLFGACLDQLLDRL 132
DG+ AR+ ++ G LD L D+L
Sbjct 111 DGYIARKYGLKTIAGTILDPLADKL 135
> ath:AT2G39290 PGP1; PGP1 (PHOSPHATIDYLGLYCEROLPHOSPHATE SYNTHASE
1); CDP-alcohol phosphatidyltransferase/ CDP-diacylglycerol-glycerol-3-phosphate
3-phosphatidyltransferase; K00995
CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase
[EC:2.7.8.5]
Length=296
Score = 29.3 bits (64), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 97 CYAASQLLDAADGHTARRLNQVSLFGACLDQLLDRL 132
+ A+ + D DG+ AR++ S FGA LD + D+L
Sbjct 140 IFIAAAITDWLDGYLARKMRLGSAFGAFLDPVADKL 175
> bbo:BBOV_IV011360 23.m05792; ethanolamine phosphatidyltransferase
(EC:2.7.8.1); K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=401
Score = 29.3 bits (64), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 35/84 (41%), Gaps = 14/84 (16%)
Query 66 VLLYVPNLIGYLRLLLLLVAALVGSQWLCLL---CYAASQLLDAADGHTARRLNQVSLFG 122
+L Y+PNL + +WL L+ C LD DG AR+L S G
Sbjct 69 ILAYIPNL-----------ESSKTPEWLPLMIAGCILLYMTLDGIDGKQARKLGMSSPVG 117
Query 123 ACLDQLLDRLSTCLFYLLNAAVCP 146
LD +D + + + + A+ P
Sbjct 118 QLLDHGVDAVVSVFYPYMCFAIYP 141
> eco:b1408 ynbA, ECK1401, JW1405; inner membrane protein
Length=201
Score = 28.9 bits (63), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 104 LDAADGHTARRLNQVSLFGACLDQLLDRLSTCLFYL 139
L+A DG AR NQ + GA L++ D +S YL
Sbjct 68 LNALDGMLARECNQQTRLGAILNETGDVISDIALYL 103
Lambda K H
0.328 0.141 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4535951560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40