bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
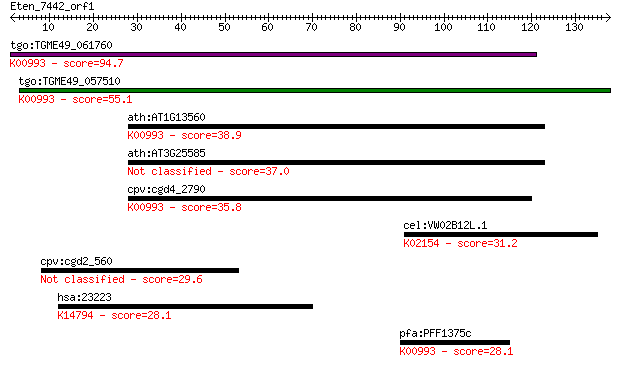
Query= Eten_7442_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_061760 ethanolaminephosphotransferase, putative (EC... 94.7 8e-20
tgo:TGME49_057510 aminoalcoholphosphotransferase, putative (EC... 55.1 6e-08
ath:AT1G13560 AAPT1; AAPT1 (AMINOALCOHOLPHOSPHOTRANSFERASE 1);... 38.9 0.004
ath:AT3G25585 AAPT2; AAPT2 (AMINOALCOHOLPHOSPHOTRANSFERASE); p... 37.0 0.016
cpv:cgd4_2790 ethanolaminephosphotransferase (ETHPT) 9 transme... 35.8 0.043
cel:VW02B12L.1 vha-6; Vacuolar H ATPase family member (vha-6);... 31.2 0.93
cpv:cgd2_560 hypothetical protein 29.6 3.0
hsa:23223 RRP12, DKFZp762P1116, FLJ20231, KIAA0690; ribosomal ... 28.1 7.5
pfa:PFF1375c ethanolaminephosphotransferase, putative (EC:2.7.... 28.1 9.3
> tgo:TGME49_061760 ethanolaminephosphotransferase, putative (EC:2.7.8.1);
K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=449
Score = 94.7 bits (234), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 52/122 (42%), Positives = 72/122 (59%), Gaps = 2/122 (1%)
Query 1 QVVMFLVYIFVQGLVYMHGLV--WRPVLNPYLIYTAICSTYAILLLRLCLSATCRFPYKL 58
QV F+ Y+ +QG ++ L W +P LIY + ++Y+ILLLR+CLSATCRFP+KL
Sbjct 266 QVAGFVGYMALQGALWHTCLEGPWEARTSPGLIYFTVTTSYSILLLRICLSATCRFPFKL 325
Query 59 VQWPMLPLSAAAAALLLGDLTADQELWAVLGVCIFNCIYLADFVYTVVSDVCDGLNTRCF 118
V P +P AAA ++ + + V ++N YL DF+YT VSDVC L CF
Sbjct 326 VNAPAIPFFLAAAGIVTSPWCREHRYALLTLVSLWNVAYLTDFLYTSVSDVCSSLEISCF 385
Query 119 SV 120
V
Sbjct 386 RV 387
> tgo:TGME49_057510 aminoalcoholphosphotransferase, putative (EC:2.7.8.1);
K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=467
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 43/162 (26%), Positives = 70/162 (43%), Gaps = 36/162 (22%)
Query 3 VMFLVYIFVQGLVYMHGLVWRPVLNPYLIYTAICSTYAILLLRLCLSATCRFPYKLVQWP 62
V+F F GL+ H P + Y+ I +I++LR+ ++ATCR + VQWP
Sbjct 276 VLFQFAFFCSGLMQTH---------PAVCYSLIIINSSIVVLRMNIAATCRLRFYPVQWP 326
Query 63 MLPLSAAA--------------AALLLGDLTADQEL--------WAVLGVCIFNCIYLAD 100
LP A + AA+L L+ Q+ W + V +++ YL D
Sbjct 327 ALPFYATSLYFYLACPNPSSPLAAVLSSPLSEAQQAYQQKHVIPWVLTAVSLWSVFYLYD 386
Query 101 FVYTVVSDVCDGLNTRCFSV-----PKFGTAQTPSEQPKKTD 137
F+ + ++ +C L+ CF V G + PS K +
Sbjct 387 FLLSTITAICSHLDITCFCVNAKEEKGKGRGRGPSAGSPKLE 428
> ath:AT1G13560 AAPT1; AAPT1 (AMINOALCOHOLPHOSPHOTRANSFERASE 1);
phosphatidyltransferase/ phosphotransferase, for other substituted
phosphate groups (EC:2.7.8.1); K00993 ethanolaminephosphotransferase
[EC:2.7.8.1]
Length=389
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 28/102 (27%), Positives = 43/102 (42%), Gaps = 7/102 (6%)
Query 28 PYLIYTAICSTYAILLLRLCLSATCRFPYKLVQ---WPMLPLSAAAAALLLGDLTAD--- 81
P+L+ + L+ R+ L+ C P L +L L A A L L A
Sbjct 285 PHLVVLGTGLAFGFLVGRMILAHLCDEPKGLKTNMCMSLLYLPFALANALTARLNAGVPL 344
Query 82 -QELWAVLGVCIFNCIYLADFVYTVVSDVCDGLNTRCFSVPK 122
ELW +LG CIF F +V+ ++ + L CF + +
Sbjct 345 VDELWVLLGYCIFTVSLYLHFATSVIHEITEALGIYCFRITR 386
> ath:AT3G25585 AAPT2; AAPT2 (AMINOALCOHOLPHOSPHOTRANSFERASE);
phosphatidyltransferase/ phosphotransferase, for other substituted
phosphate groups
Length=337
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 43/102 (42%), Gaps = 7/102 (6%)
Query 28 PYLIYTAICSTYAILLLRLCLSATCRFPYKL-----VQWPMLPLSAAAA--ALLLGDLTA 80
P+L+ + L+ R+ L+ C P L + LP + A A A L +
Sbjct 233 PHLVVLGTGLAFGFLVGRMILAHLCDEPKGLKTNMCMSLLYLPFALANALTARLNDGVPL 292
Query 81 DQELWAVLGVCIFNCIYLADFVYTVVSDVCDGLNTRCFSVPK 122
E W +LG CIF A F +V+ ++ L CF + +
Sbjct 293 VDEFWVLLGYCIFTLSLYAHFATSVIHEITTALGIYCFRITR 334
> cpv:cgd4_2790 ethanolaminephosphotransferase (ETHPT) 9 transmembrane
domain protein involved in lipid ; K00993 ethanolaminephosphotransferase
[EC:2.7.8.1]
Length=428
Score = 35.8 bits (81), Expect = 0.043, Method: Composition-based stats.
Identities = 24/96 (25%), Positives = 46/96 (47%), Gaps = 5/96 (5%)
Query 28 PYLIYTAICSTYAILLLRLCLSATCRFPYKLVQWPMLPLSAAAAALLLGDLT----ADQE 83
P + + ++ +I+ LR+ +S+ V WP LP ++ LL G D +
Sbjct 286 PIMTMFILTTSSSIVALRMNVSSFTLEELPYVHWPALPFYTSSVFLLFGRNIFGKFFDSQ 345
Query 84 LWAVLGVCIFNCIYLADFVYTVVSDVCDGLNTRCFS 119
+ +L V ++N IY D+V +++ +C L+ S
Sbjct 346 I-ILLIVALWNVIYTIDYVSIIINSICHHLSISLIS 380
> cel:VW02B12L.1 vha-6; Vacuolar H ATPase family member (vha-6);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=865
Score = 31.2 bits (69), Expect = 0.93, Method: Composition-based stats.
Identities = 12/44 (27%), Positives = 23/44 (52%), Gaps = 5/44 (11%)
Query 91 CIFNCIYLADFVYTVVSDVCDGLNTRCFSVPKFGTAQTPSEQPK 134
C+F + + + V +CDG +C++VP + P+E+ K
Sbjct 229 CVFILFFSGEQLRAKVKKICDGFQAKCYTVP-----ENPAERTK 267
> cpv:cgd2_560 hypothetical protein
Length=2338
Score = 29.6 bits (65), Expect = 3.0, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 20/45 (44%), Gaps = 0/45 (0%)
Query 8 YIFVQGLVYMHGLVWRPVLNPYLIYTAICSTYAILLLRLCLSATC 52
Y+ + M V +LNPYLI T Y LLLR S C
Sbjct 983 YVDEDNSILMENNVQTSLLNPYLILTGSFPIYNTLLLRSLYSIMC 1027
> hsa:23223 RRP12, DKFZp762P1116, FLJ20231, KIAA0690; ribosomal
RNA processing 12 homolog (S. cerevisiae); K14794 ribosomal
RNA-processing protein 12
Length=1236
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 24/58 (41%), Gaps = 4/58 (6%)
Query 12 QGLVYMHGLVWRPVLNPYLIYTAICSTYAILLLRLCLSATCRFPYKLVQWPMLPLSAA 69
+GL Y W VL ++ C A ++R CL + C L P P +AA
Sbjct 416 EGLTYKFHAAWSSVLQLLCVFFEACGRQAHPVMRKCLQSLC----DLRLSPHFPHTAA 469
> pfa:PFF1375c ethanolaminephosphotransferase, putative (EC:2.7.8.1);
K00993 ethanolaminephosphotransferase [EC:2.7.8.1]
Length=391
Score = 28.1 bits (61), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 9/25 (36%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 90 VCIFNCIYLADFVYTVVSDVCDGLN 114
+ F IYL D+ +T+++++C LN
Sbjct 355 ILFFGIIYLLDYAHTIITNICKELN 379
Lambda K H
0.330 0.142 0.468
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40