bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
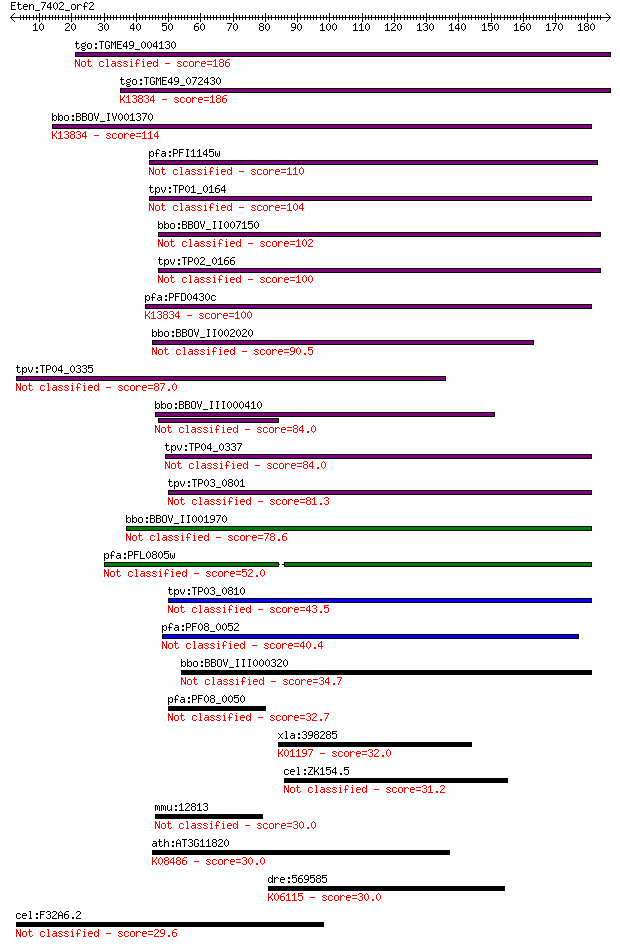
Query= Eten_7402_orf2
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_004130 membrane-attack complex / perforin domain-co... 186 4e-47
tgo:TGME49_072430 membrane-attack complex / perforin domain-co... 186 5e-47
bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing pr... 114 2e-25
pfa:PFI1145w MAC/Perforin, putative 110 3e-24
tpv:TP01_0164 hypothetical protein 104 2e-22
bbo:BBOV_II007150 18.m06592; mac/perforin domain containing pr... 102 7e-22
tpv:TP02_0166 hypothetical protein 100 3e-21
pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microne... 100 4e-21
bbo:BBOV_II002020 18.m06160; mac/perforin domain containing pr... 90.5 3e-18
tpv:TP04_0335 hypothetical protein 87.0 4e-17
bbo:BBOV_III000410 hypothetical protein 84.0 3e-16
tpv:TP04_0337 hypothetical protein 84.0 3e-16
tpv:TP03_0801 hypothetical protein 81.3 2e-15
bbo:BBOV_II001970 18.m09950; mac/perforin domain containing me... 78.6 1e-14
pfa:PFL0805w MAC/Perforin, putative 52.0 1e-06
tpv:TP03_0810 hypothetical protein 43.5 5e-04
pfa:PF08_0052 perforin like protein 5 40.4
bbo:BBOV_III000320 17.m10445; hypothetical protein 34.7 0.18
pfa:PF08_0050 MAC/Perforin, putative 32.7 0.76
xla:398285 spam1; sperm adhesion molecule 1 (PH-20 hyaluronida... 32.0 1.2
cel:ZK154.5 hypothetical protein 31.2 2.2
mmu:12813 Col10a1, Col10a-1; collagen, type X, alpha 1 30.0
ath:AT3G11820 SYP121; SYP121 (SYNTAXIN OF PLANTS 121); SNAP re... 30.0 4.8
dre:569585 Spectrin beta chain, putative-like; K06115 spectrin... 30.0 5.1
cel:F32A6.2 ift-81; IFT (Chlamydomonas IntraFlagellar Transpor... 29.6 5.4
> tgo:TGME49_004130 membrane-attack complex / perforin domain-containing
protein
Length=1054
Score = 186 bits (472), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 83/170 (48%), Positives = 120/170 (70%), Gaps = 4/170 (2%)
Query 21 RLRE----MELSHKKASIQYLYSTVKTVSPGLNYLGAGYDGVKGNPLGDPNLMGDPGLRV 76
R+RE E + A + +Y+ P +NYLGAGYD V+GNP+GDP+ MGDPG+R
Sbjct 339 RMRENRRIAEENRAAAPLSAVYTKATKTVPAINYLGAGYDHVRGNPVGDPSSMGDPGIRP 398
Query 77 PIIQFTFLQDAEGVSRDLKELQPLGAYSRPFVACKQSETLSEVATLADYLQELAADAAVA 136
P+++FT+ Q+ +GVS DL LQPLG Y R +VAC+QSET+SE++ L+DY EL+ DA++
Sbjct 399 PVLRFTYAQNEDGVSNDLTVLQPLGGYVRQYVACRQSETISELSNLSDYQNELSVDASLQ 458
Query 137 AGDTLGFNAFSASAAYREQARKTVQKKSHNFILKTYCLRYEDGLTQSDDF 186
GD +G N+FSAS YR+ A++ +K + ++LK YC+RYE G+ QS+ F
Sbjct 459 GGDPIGLNSFSASTGYRDFAKEVSKKDTRTYMLKNYCMRYEAGVAQSNHF 508
> tgo:TGME49_072430 membrane-attack complex / perforin domain-containing
protein ; K13834 sporozoite microneme protein 2
Length=854
Score = 186 bits (471), Expect = 5e-47, Method: Compositional matrix adjust.
Identities = 81/152 (53%), Positives = 112/152 (73%), Gaps = 0/152 (0%)
Query 35 QYLYSTVKTVSPGLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDL 94
+ +Y+ +P N+LG GYD +KGNP+GDP++M DPGLR PII F+F QD +GV+ DL
Sbjct 220 ESMYTRAVETAPATNFLGVGYDSIKGNPIGDPDMMVDPGLRSPIIVFSFQQDPDGVTNDL 279
Query 95 KELQPLGAYSRPFVACKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYRE 154
LQPLGA++RPF AC+QSE ++E+ TL+DY + L+ DAA+ GD+LG N+FS S Y+E
Sbjct 280 NYLQPLGAFTRPFSACRQSENVNELDTLSDYQKVLSVDAALHGGDSLGINSFSGSTGYKE 339
Query 155 QARKTVQKKSHNFILKTYCLRYEDGLTQSDDF 186
A+ K + +F+LKTYC+RYE GL Q+D F
Sbjct 340 FAQDVSSKANKSFMLKTYCIRYEAGLAQTDSF 371
> bbo:BBOV_IV001370 21.m02755; MAC/perforin domain containing
protein; K13834 sporozoite microneme protein 2
Length=978
Score = 114 bits (285), Expect = 2e-25, Method: Composition-based stats.
Identities = 69/181 (38%), Positives = 107/181 (59%), Gaps = 22/181 (12%)
Query 14 RSGEQERRLREMELSHKKASIQYLYSTVKTVS-------PGL----NYLGAGYDGVKGNP 62
R E+ RRL+E S ++ + ++ V+ PGL YLG+GYD + GNP
Sbjct 286 RLSEENRRLKE---SVDRSYYERMFRPETDVTDAEGRLNPGLAAAMRYLGSGYDIIYGNP 342
Query 63 LGDPNLMGDPGLRVPIIQFTFLQ---DAEGVSRDLKELQPLGAYSRPFVACKQSETLSEV 119
LGDP +M DPG R P+++ + + + +G ++KE P G + RP ++C+QSE++ +
Sbjct 343 LGDPVIMVDPGYRHPVLRLDWSEKYYNNDGA--NMKE--PKGGWIRPELSCRQSESVDHI 398
Query 120 ATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQARKTVQKKSHNFILKTYCLRYEDG 179
T+ DY +EL+ DA ++A L F +FSASA Y+ R ++ N+ILKTYCLRY G
Sbjct 399 NTMDDYKKELSVDAKMSADMPLYF-SFSASAGYKNMVRTLATNETKNYILKTYCLRYVAG 457
Query 180 L 180
+
Sbjct 458 I 458
> pfa:PFI1145w MAC/Perforin, putative
Length=821
Score = 110 bits (274), Expect = 3e-24, Method: Composition-based stats.
Identities = 54/139 (38%), Positives = 83/139 (59%), Gaps = 1/139 (0%)
Query 44 VSPGLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAY 103
V G YLG GYD + GNP+GDP L DPG R II+ T+ + E + + P G++
Sbjct 235 VIQGTEYLGVGYDFIFGNPIGDPFLKVDPGYRDSIIKLTYPKSDEDYPDNYMNINPNGSF 294
Query 104 SRPFVACKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQARKTVQKK 163
R ++C +SE SE++T+++Y +EL+ DA++ A L F +FSAS Y+ + + K
Sbjct 295 VRNEISCNRSEKESEISTMSEYTKELSVDASIGASYGL-FGSFSASTGYKSVSNTISKNK 353
Query 164 SHNFILKTYCLRYEDGLTQ 182
F+LK+YC +Y L+Q
Sbjct 354 FRMFMLKSYCFKYVASLSQ 372
> tpv:TP01_0164 hypothetical protein
Length=1182
Score = 104 bits (259), Expect = 2e-22, Method: Composition-based stats.
Identities = 56/138 (40%), Positives = 85/138 (61%), Gaps = 4/138 (2%)
Query 44 VSPGLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKEL-QPLGA 102
++ + YLG+GYD + GNPLGDP +M D G R P+I+ + + E +++D L +P G+
Sbjct 583 LAASMRYLGSGYDIIFGNPLGDPVVMMDQGYRNPVIRLNW--EDEYLNKDGANLKEPRGS 640
Query 103 YSRPFVACKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQARKTVQK 162
+ RP +C+QSET+ V T+ D+ +EL+ DA + G F +FSAS Y+ + T
Sbjct 641 WIRPEYSCRQSETIDHVNTVDDFKKELSVDAQASYGIPY-FFSFSASTGYKNFVKSTATN 699
Query 163 KSHNFILKTYCLRYEDGL 180
K +I KTYCLRY G+
Sbjct 700 KVRTYITKTYCLRYVGGI 717
> bbo:BBOV_II007150 18.m06592; mac/perforin domain containing
protein
Length=752
Score = 102 bits (254), Expect = 7e-22, Method: Composition-based stats.
Identities = 55/140 (39%), Positives = 81/140 (57%), Gaps = 6/140 (4%)
Query 47 GLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAYSRP 106
GL YLGAGYD +KGNP+GD ++ DPG R ++Q + DAEG+S +QP GA+ RP
Sbjct 421 GLEYLGAGYDLLKGNPMGDTIILLDPGYRASVVQMHWRDDAEGLSNSRHFIQPKGAWVRP 480
Query 107 FVACKQSETLSEVATLADYLQELAADAAVAA---GDTLGFNAFSASAAYREQARKTVQKK 163
+ +C + ET+SEVA L+ADA+V+A GD F+AS Y + K
Sbjct 481 YTSCHKGETISEVAKTQSLDNVLSADASVSASLPGDKF---KFAASVNYNNIKKAYDSKG 537
Query 164 SHNFILKTYCLRYEDGLTQS 183
+ ++ ++YC + G+ S
Sbjct 538 VNTYVSRSYCFNFVAGIPMS 557
> tpv:TP02_0166 hypothetical protein
Length=812
Score = 100 bits (249), Expect = 3e-21, Method: Composition-based stats.
Identities = 52/137 (37%), Positives = 83/137 (60%), Gaps = 3/137 (2%)
Query 47 GLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAYSRP 106
GL YLGAGYD ++GNPLGD + DPG + +IQ + ++ E +S L+ LQP+G + RP
Sbjct 347 GLEYLGAGYDLIRGNPLGDSVTLLDPGYKSSVIQMHWSRNIENISNSLRFLQPVGGWIRP 406
Query 107 FVACKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQARKTVQKKSHN 166
+ +C + ++E+ + L+ L+ADA+V+ FSASA +++ + KS
Sbjct 407 YSSCHK---VTEINSCKSLLKSLSADASVSLSLPGDVFKFSASAKFKKLQDVSKSGKSKM 463
Query 167 FILKTYCLRYEDGLTQS 183
FI K+YC +Y G++ S
Sbjct 464 FINKSYCFKYVAGISTS 480
> pfa:PFD0430c MAC/Perforin, putative; K13834 sporozoite microneme
protein 2
Length=842
Score = 100 bits (248), Expect = 4e-21, Method: Composition-based stats.
Identities = 48/138 (34%), Positives = 82/138 (59%), Gaps = 1/138 (0%)
Query 43 TVSPGLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGA 102
+V PGL ++G GY+ + GNPLG+ + + DPG R I + EG++ DL LQP+
Sbjct 239 SVFPGLYFIGIGYNLLFGNPLGEADSLIDPGYRAQIYLMEWALSKEGIANDLSTLQPVNG 298
Query 103 YSRPFVACKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQARKTVQK 162
+ R AC + E+++E ++++DY + L+A+A V +G G +FSAS Y + ++
Sbjct 299 WIRKENACSRVESITECSSISDYTKSLSAEAKV-SGSYWGIASFSASTGYSSFLHEVTKR 357
Query 163 KSHNFILKTYCLRYEDGL 180
F++K+ C++Y GL
Sbjct 358 SKKTFLVKSNCVKYTIGL 375
> bbo:BBOV_II002020 18.m06160; mac/perforin domain containing
protein
Length=420
Score = 90.5 bits (223), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 46/118 (38%), Positives = 68/118 (57%), Gaps = 2/118 (1%)
Query 45 SPGLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAYS 104
+PG++YLG GYD +KGN +G + DPG R PII F + + AEG S L + PL +
Sbjct 75 TPGMDYLGIGYDAIKGNTMGGEESLLDPGYRAPIINFNWRKSAEGYSPSLNAVYPLYGWV 134
Query 105 RPFVACKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQARKTVQK 162
RP +C +S + E+ L + + +A+A++ GD AFSASA Y+ + K K
Sbjct 135 RPVYSCGRSSKIQEIENLDELKKVFSANASI-KGDIPAV-AFSASAKYKNASEKLAHK 190
> tpv:TP04_0335 hypothetical protein
Length=441
Score = 87.0 bits (214), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 48/133 (36%), Positives = 72/133 (54%), Gaps = 3/133 (2%)
Query 3 QRRQHQQQKNWRSGEQERRLREMELSHKKASIQYLYSTVKTVSPGLNYLGAGYDGVKGNP 62
+ + H + + GE + E+E + SI + T K V GL YLGAGYD VK N
Sbjct 31 ETKIHSLFEKGKGGEANKSKSEVEPIVENESINF--KTTKIVV-GLEYLGAGYDIVKANT 87
Query 63 LGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAYSRPFVACKQSETLSEVATL 122
+GD + D G R P+I FT+ Q GV+ L LQP+G + RP V+C +SE ++E+ ++
Sbjct 88 MGDADQAEDLGYRAPVIDFTWAQTDVGVTNSLDSLQPVGGWVRPKVSCGESENVTEIESI 147
Query 123 ADYLQELAADAAV 135
+ +D V
Sbjct 148 SKLKDVTESDVGV 160
> bbo:BBOV_III000410 hypothetical protein
Length=1272
Score = 84.0 bits (206), Expect = 3e-16, Method: Composition-based stats.
Identities = 43/105 (40%), Positives = 58/105 (55%), Gaps = 0/105 (0%)
Query 46 PGLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAYSR 105
P + YLG GYD + GNPL D + DPG R PII FT +DLK GA+ R
Sbjct 942 PAIEYLGCGYDILYGNPLADDGTLVDPGYRNPIISFTLAHHKSKGKKDLKYANIPGAWIR 1001
Query 106 PFVACKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASA 150
P VAC++S+ S V +++DY + L+ D+ V G F+ SA
Sbjct 1002 PLVACQRSDETSIVKSISDYQKALSVDSEVGIGTVDESAKFALSA 1046
Score = 38.1 bits (87), Expect = 0.016, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 47 GLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTF 83
GL YLG GYD K P GD D G P++ F +
Sbjct 136 GLEYLGCGYDSTKSMPFGDEESFLDSGYTQPVVNFQW 172
> tpv:TP04_0337 hypothetical protein
Length=498
Score = 84.0 bits (206), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 42/132 (31%), Positives = 72/132 (54%), Gaps = 2/132 (1%)
Query 49 NYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAYSRPFV 108
+YLG GYD + N +G + + DPG R PII+F + +++EG S L L P+G + RP
Sbjct 16 DYLGVGYDSIYANSVGSDSTLLDPGYRAPIIEFAWRKNSEGYSPTLGSLHPVGGWVRPVF 75
Query 109 ACKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQARKTVQKKSHNFI 168
+C +S ++E++ L + L+A + GD + N+F+ S Y+ K+ +
Sbjct 76 SCSRSTKINEISNLEELKDSLSASTKL-NGD-IPENSFTGSLEYKNALMNFKSKRQKIYN 133
Query 169 LKTYCLRYEDGL 180
C+RY+ G+
Sbjct 134 KTEQCVRYQVGI 145
> tpv:TP03_0801 hypothetical protein
Length=353
Score = 81.3 bits (199), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 65/131 (49%), Gaps = 0/131 (0%)
Query 50 YLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAYSRPFVA 109
YLG GYD + GNPL DP+L+ DPG R PII ++ + E + + + A+ RP +
Sbjct 26 YLGCGYDILFGNPLSDPDLLVDPGFRDPIISYSIMFKKEKLFKKISYSNITNAWIRPLIE 85
Query 110 CKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQARKTVQKKSHNFIL 169
CK+S + S V ++ Y ++ D+ + FS S Y E + + ++
Sbjct 86 CKRSNSRSVVDSMEKYKDIISVDSDIGVSSIDESAKFSLSTNYSEISDLLKNNDNKLYVD 145
Query 170 KTYCLRYEDGL 180
K+YC E L
Sbjct 146 KSYCFLLEAAL 156
> bbo:BBOV_II001970 18.m09950; mac/perforin domain containing
membrane protein
Length=559
Score = 78.6 bits (192), Expect = 1e-14, Method: Composition-based stats.
Identities = 54/147 (36%), Positives = 76/147 (51%), Gaps = 8/147 (5%)
Query 37 LYSTVKTVSPGLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKE 96
++S + VS GL YLG+GYD VK + L N D G R PI+ F + + GV+ LK
Sbjct 49 IFSADRIVS-GLEYLGSGYDAVKASGLVSINNGDDLGHRSPIVDFYWAKSDVGVTNSLKW 107
Query 97 LQPLGAYSRPFVACKQSETL---SEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYR 153
LQPLG + RP AC +SET+ S+ +T + Q A+++G LG A A Y
Sbjct 108 LQPLGGWVRPITACGESETVTVGSQQSTNEESTQFDFGGFAMSSG--LGSGAL--RAGYT 163
Query 154 EQARKTVQKKSHNFILKTYCLRYEDGL 180
+ + KT S + YC Y G+
Sbjct 164 DISGKTQHISSKQYTNSYYCFTYAAGM 190
> pfa:PFL0805w MAC/Perforin, putative
Length=1073
Score = 52.0 bits (123), Expect = 1e-06, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 30 KKASIQYLYSTVKTVSPGLNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTF 83
+K +++ +Y+ K L YLG GYD + GNP GDP L DPG R P++Q
Sbjct 384 RKENLKEVYNDKKNNYLSLKYLGLGYDIIMGNPEGDPTLNVDPGFRGPVLQINL 437
Score = 50.4 bits (119), Expect = 3e-06, Method: Composition-based stats.
Identities = 30/100 (30%), Positives = 54/100 (54%), Gaps = 6/100 (6%)
Query 86 DAEGVSRDLK-ELQPLGAYSRPFV----ACKQSETLSEVATLADYLQELAADAAVAAGDT 140
D + S++ K ++Q +P+V +C QS+ + E+ L Y EL +D V+ +
Sbjct 500 DGDKRSKERKSKIQTTNESMKPWVIPEHSCSQSKNVEEIRNLEQYKLELLSDVKVSTPSS 559
Query 141 LGFNAFSASAAYREQARKTVQKKSHNFILKTYCLRYEDGL 180
+ +FSASA ++ +K + + F++K YCLRY G+
Sbjct 560 FPY-SFSASAEFKNALKKLKVQNNVIFLMKIYCLRYYTGI 598
> tpv:TP03_0810 hypothetical protein
Length=348
Score = 43.5 bits (101), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 61/144 (42%), Gaps = 32/144 (22%)
Query 50 YLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKEL---QPLGAYSRP 106
++G G+D V+GNPL N + G R PII + ++RD+ + + G + R
Sbjct 38 HVGFGFDLVEGNPLDSFNDLNTFGFRSPIIVQPY------ITRDIGNIIIKRNNGIWVRK 91
Query 107 FVACKQSETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQARKTVQK---- 162
C Q+ ++ +D ++EL FN FS + + E+ K
Sbjct 92 SNNCTQNYEPRDIERGSDLVREL-------------FNDFSLDSPFSEELWNRNSKGLGL 138
Query 163 ------KSHNFILKTYCLRYEDGL 180
KS I+K YC YE GL
Sbjct 139 NDITFNKSKFKIVKCYCSLYESGL 162
> pfa:PF08_0052 perforin like protein 5
Length=676
Score = 40.4 bits (93), Expect = 0.004, Method: Composition-based stats.
Identities = 34/131 (25%), Positives = 58/131 (44%), Gaps = 9/131 (6%)
Query 48 LNYLGAGYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAYSRPF 107
L YLG YD +KGNP GDP + D G R +++ L + D+ + + +G S+
Sbjct 47 LKYLGMSYDIIKGNPWGDPIYVIDLGYRRNVLKKRKLNSDNNIKDDIVKFK-VGEASK-- 103
Query 108 VACKQSETLSEVATLADYLQELAADAAVAA--GDTLGFNAFSASAAYREQARKTVQKKSH 165
+ C + + + L D +E +V++ D FN + Y + K + +
Sbjct 104 IKCIDTIKENVIDNLCDINKEYERSYSVSSINDDIHPFN----DSNYYKMLVKRINRGDS 159
Query 166 NFILKTYCLRY 176
I K C +Y
Sbjct 160 IIIEKKLCSKY 170
> bbo:BBOV_III000320 17.m10445; hypothetical protein
Length=512
Score = 34.7 bits (78), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 38/130 (29%), Positives = 56/130 (43%), Gaps = 13/130 (10%)
Query 54 GYDGVKGNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRD-LKELQPLGAYSRPFVACKQ 112
GYD V GNP G D G R PI L+ +S D K + G + R C
Sbjct 2 GYDAVLGNPFGSLGQDKDSGYRNPI-----LETHVTISGDKTKSSEQNGLWVRELSTCWI 56
Query 113 SETLSEVATLADYLQELAADAAVAAGDTLGFNAFSASAAYREQA-RKTVQKKSHNF-ILK 170
S+T +V + ++EL + V + N+ SA+ A K K + N+ I K
Sbjct 57 SDTHDDVGD-DELVRELQNEFTVEGSE----NSELLSASINSMADDKPTSKHTVNYRIAK 111
Query 171 TYCLRYEDGL 180
++C E G+
Sbjct 112 SFCAIRESGI 121
> pfa:PF08_0050 MAC/Perforin, putative
Length=654
Score = 32.7 bits (73), Expect = 0.76, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 50 YLGAGYDGVKGNPLGDPNLMGDPGLRVPII 79
YLG GYD + G PL + L+ DPG + II
Sbjct 43 YLGKGYDILFGYPLPNNELIDDPGFKEVII 72
> xla:398285 spam1; sperm adhesion molecule 1 (PH-20 hyaluronidase,
zona pellucida binding) (EC:3.2.1.35); K01197 hyaluronoglucosaminidase
[EC:3.2.1.35]
Length=512
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 32/63 (50%), Gaps = 4/63 (6%)
Query 84 LQDAEGVSRDLKELQPLGAYSRPFVACKQSETLSEVATLADYLQELA---ADAAVAAGDT 140
+Q+A +S K P+ Y+RP + E LSEV LA+ + E+A AD V GD
Sbjct 287 IQEARRLSTFSKYAVPIYVYTRPVFTNRPDEFLSEV-DLANVIGEIAALGADGFVMWGDV 345
Query 141 LGF 143
+
Sbjct 346 INM 348
> cel:ZK154.5 hypothetical protein
Length=1105
Score = 31.2 bits (69), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 19/76 (25%), Positives = 41/76 (53%), Gaps = 8/76 (10%)
Query 86 DAEGVSRDLKELQPLGAYSRPFV-------ACKQSETLSEVATLADYLQELAADAAVAAG 138
D + S ++ ++P+ A++RP + SE+ +V T +Y++E + +A G
Sbjct 952 DLKTTSEEIVYMRPITAHTRPSARDLFEKNKSEPSESRYKVKTREEYMKENDEEEELA-G 1010
Query 139 DTLGFNAFSASAAYRE 154
+T+ F F+ + AY++
Sbjct 1011 ETVSFEYFTENHAYQD 1026
> mmu:12813 Col10a1, Col10a-1; collagen, type X, alpha 1
Length=680
Score = 30.0 bits (66), Expect = 4.7, Method: Composition-based stats.
Identities = 20/43 (46%), Positives = 24/43 (55%), Gaps = 10/43 (23%)
Query 46 PGLN----YLG-AGYDGVKGNP-----LGDPNLMGDPGLRVPI 78
PGL+ Y G G +G KGNP GDP + G PGLR P+
Sbjct 382 PGLDGKTGYPGEPGLNGPKGNPGLPGQKGDPGVGGTPGLRGPV 424
> ath:AT3G11820 SYP121; SYP121 (SYNTAXIN OF PLANTS 121); SNAP
receptor/ protein anchor; K08486 syntaxin 1B/2/3
Length=346
Score = 30.0 bits (66), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 42/94 (44%), Gaps = 15/94 (15%)
Query 45 SPGLNYLGAGYDGVK-GNPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKELQPLGAY 103
SP + G G DGV+ NP G G+ + F +D E V +LKEL L
Sbjct 18 SPRRDVAGGG-DGVQMANPAGSTG-----GVNLD----KFFEDVESVKEELKELDRL--- 64
Query 104 SRPFVAC-KQSETLSEVATLADYLQELAADAAVA 136
+ +C +QS+TL + D ++ D VA
Sbjct 65 NETLSSCHEQSKTLHNAKAVKDLRSKMDGDVGVA 98
> dre:569585 Spectrin beta chain, putative-like; K06115 spectrin
beta
Length=4136
Score = 30.0 bits (66), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 9/73 (12%)
Query 81 FTFLQDAEGVSRDLKELQPLGAYSRPFVACKQSETLSEVATLADYLQELAADAAVAAGDT 140
F L+D E +S DLKELQ S + L +V L Q+L ++ G+T
Sbjct 502 FAVLRDIELISMDLKELQAQADSS------DMGKQLPDVVALLQK-QDLIDTQIISLGET 554
Query 141 LGFNAFSASAAYR 153
L NA S+S+A +
Sbjct 555 L--NAISSSSALK 565
> cel:F32A6.2 ift-81; IFT (Chlamydomonas IntraFlagellar Transport)
homolog family member (ift-81)
Length=590
Score = 29.6 bits (65), Expect = 5.4, Method: Composition-based stats.
Identities = 25/98 (25%), Positives = 45/98 (45%), Gaps = 10/98 (10%)
Query 3 QRRQHQQQKNWRSGEQERRLREMELSHKKASIQYLYSTVKTVSPGLNYLGAGYDGVKG-- 60
Q R++ +Q +S EQ+++ +E+EL H + ++ L T+ VS N L + + G
Sbjct 348 QYRKYLEQYRVKSEEQKKKRKEIELMHTEVAV--LKRTIDIVSKRYNKLETHIESIGGEI 405
Query 61 -NPLGDPNLMGDPGLRVPIIQFTFLQDAEGVSRDLKEL 97
L P P P +AE + D+K++
Sbjct 406 VEVLNVPTKYDRPKTAAPQT-----NNAEDIKSDMKDM 438
Lambda K H
0.316 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40