bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7386_orf1
Length=129
Score E
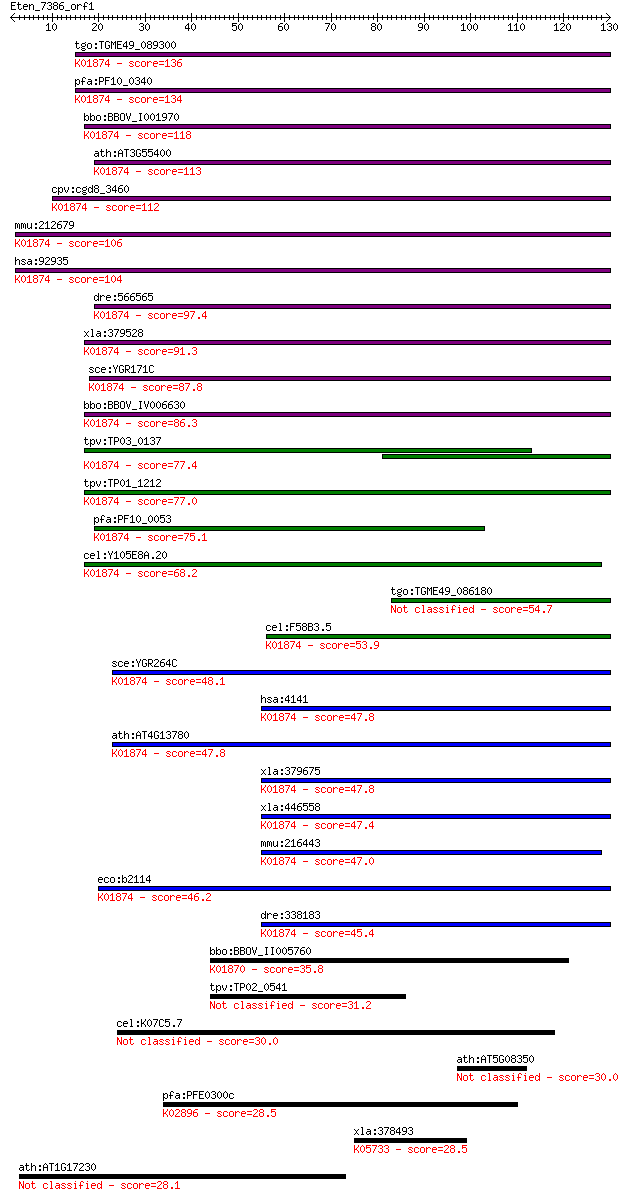
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_089300 methionyl-tRNA synthetase, putative (EC:6.1.... 136 2e-32
pfa:PF10_0340 methionine-tRNA ligase, putative; K01874 methion... 134 8e-32
bbo:BBOV_I001970 19.m02057; methionyl-tRNA synthetase (EC:6.1.... 118 5e-27
ath:AT3G55400 OVA1; OVA1 (OVULE ABORTION 1); ATP binding / ami... 113 1e-25
cpv:cgd8_3460 methionyl-tRNA synthetase of possible bacterial ... 112 2e-25
mmu:212679 Mars2, C730026E21Rik, MetRS; methionine-tRNA synthe... 106 2e-23
hsa:92935 MARS2, MetRS, mtMetRS; methionyl-tRNA synthetase 2, ... 104 8e-23
dre:566565 Methionyl-tRNA synthetase, mitochondrial-like; K018... 97.4 1e-20
xla:379528 mars2, MGC68850, metrs; methionyl-tRNA synthetase 2... 91.3 7e-19
sce:YGR171C MSM1; Msm1p (EC:6.1.1.10); K01874 methionyl-tRNA s... 87.8 8e-18
bbo:BBOV_IV006630 23.m05807; methionyl-tRNA synthetase (EC:6.1... 86.3 2e-17
tpv:TP03_0137 methionyl-tRNA synthetase (EC:6.1.1.10); K01874 ... 77.4 1e-14
tpv:TP01_1212 methionyl-tRNA synthetase; K01874 methionyl-tRNA... 77.0 1e-14
pfa:PF10_0053 methionine-tRNA ligase, putative; K01874 methion... 75.1 5e-14
cel:Y105E8A.20 hypothetical protein; K01874 methionyl-tRNA syn... 68.2 7e-12
tgo:TGME49_086180 methionine tRNA synthetase, putative (EC:6.1... 54.7 7e-08
cel:F58B3.5 mrs-1; Methionyl tRNA Synthetase family member (mr... 53.9 1e-07
sce:YGR264C MES1; Mes1p (EC:6.1.1.10); K01874 methionyl-tRNA s... 48.1 7e-06
hsa:4141 MARS, FLJ35667, METRS, MRS, MTRNS; methionyl-tRNA syn... 47.8 8e-06
ath:AT4G13780 methionine--tRNA ligase, putative / methionyl-tR... 47.8 8e-06
xla:379675 mars, MGC69150; methionyl-tRNA synthetase, cytoplas... 47.8 9e-06
xla:446558 mars, MGC81271, metrs, mtrns; methionyl-tRNA synthe... 47.4 1e-05
mmu:216443 Mars, MGC90747, Metrs, Mtrns; methionine-tRNA synth... 47.0 1e-05
eco:b2114 metG, ECK2107, JW2101; methionyl-tRNA synthetase (EC... 46.2 2e-05
dre:338183 mars, id:ibd5106, zgc:66122; methionine-tRNA synthe... 45.4 4e-05
bbo:BBOV_II005760 18.m06479; tRNA synthetases class I protein ... 35.8 0.039
tpv:TP02_0541 hypothetical protein 31.2 0.88
cel:K07C5.7 ttll-15; Tubulin Tyrosine Ligase Like family membe... 30.0 1.7
ath:AT5G08350 GRAM domain-containing protein / ABA-responsive ... 30.0 2.0
pfa:PFE0300c 60S ribosomal subunit protein L24-2, putative; K0... 28.5 5.8
xla:378493 pak3, XPak3, pak3-A; p21 protein (Cdc42/Rac)-activa... 28.5 6.2
ath:AT1G17230 ATP binding / protein binding / protein kinase/ ... 28.1 6.8
> tgo:TGME49_089300 methionyl-tRNA synthetase, putative (EC:6.1.1.10);
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=976
Score = 136 bits (342), Expect = 2e-32, Method: Composition-based stats.
Identities = 59/115 (51%), Positives = 87/115 (75%), Gaps = 2/115 (1%)
Query 15 KEVAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWG 74
+++ E +YFF++SKY RL+++I NPDFI+P RR ++ +++ L DL SRT+F WG
Sbjct 427 QKMEEPSYFFRMSKYQARLVKHINENPDFIRPEERRKNILKRLEEPLLDLSCSRTTFSWG 486
Query 75 IPVPIDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLHLVGKEIVRFHTI 129
+PVP DEKHV+YVWFDALTNY + ++D + + FWPAD+H++GK+IV FHT+
Sbjct 487 VPVPGDEKHVMYVWFDALTNYYSAT-VMNDGARAS-FWPADVHIIGKDIVWFHTV 539
> pfa:PF10_0340 methionine-tRNA ligase, putative; K01874 methionyl-tRNA
synthetase [EC:6.1.1.10]
Length=889
Score = 134 bits (336), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 60/115 (52%), Positives = 88/115 (76%), Gaps = 3/115 (2%)
Query 15 KEVAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWG 74
+++ E +YFFK+SKY D+L+EYI++NPD+IQP +RNE++ +K+ L DL SRT F WG
Sbjct 374 EKMKEPSYFFKMSKYQDKLIEYIQSNPDYIQPEQKRNEILQRLKEPLADLSCSRTKFTWG 433
Query 75 IPVPIDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLHLVGKEIVRFHTI 129
I VP D KHV+YVW DAL NY + ++D+ +K K+WPA++H++GK+I FHT+
Sbjct 434 IQVPNDPKHVMYVWMDALINYYSSC-FIDEGKK--KYWPANVHIIGKDITWFHTV 485
> bbo:BBOV_I001970 19.m02057; methionyl-tRNA synthetase (EC:6.1.1.10);
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=534
Score = 118 bits (295), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 55/115 (47%), Positives = 81/115 (70%), Gaps = 3/115 (2%)
Query 17 VAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWGIP 76
+ E +YFF++SKY D L+E + ++P ++QP S R E+++ +K+ LEDL +SR +F WGIP
Sbjct 180 MKESSYFFRMSKYQDMLIEKLSSDPAYLQPDSARAEILSRLKKPLEDLSVSRCNFKWGIP 239
Query 77 VPIDEKHVIYVWFDALTNYLTPIGY--LDDPEKFNKFWPADLHLVGKEIVRFHTI 129
VP D HV+YVW DALT YL+ IG+ DD + K WP DL ++GK+I+ F +
Sbjct 240 VPDDPSHVMYVWSDALTGYLSGIGFGDFDDISQL-KLWPPDLQVIGKDIIWFFAV 293
> ath:AT3G55400 OVA1; OVA1 (OVULE ABORTION 1); ATP binding / aminoacyl-tRNA
ligase/ methionine-tRNA ligase/ nucleotide binding;
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=533
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 54/115 (46%), Positives = 72/115 (62%), Gaps = 4/115 (3%)
Query 19 EEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWGIPVP 78
E+ YFF +SKY L + + NP F+QP R NE+ ++IK GL D ISR DWGIPVP
Sbjct 223 EDNYFFALSKYQKPLEDILAQNPRFVQPSYRLNEVQSWIKSGLRDFSISRALVDWGIPVP 282
Query 79 IDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKF----WPADLHLVGKEIVRFHTI 129
D+K IYVWFDAL Y++ + + + WPA LHL+GK+I+RFH +
Sbjct 283 DDDKQTIYVWFDALLGYISALTEDNKQQNLETAVSFGWPASLHLIGKDILRFHAV 337
> cpv:cgd8_3460 methionyl-tRNA synthetase of possible bacterial
origin ; K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=581
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 59/129 (45%), Positives = 85/129 (65%), Gaps = 10/129 (7%)
Query 10 SLSLSKEVAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMI---NFIKQGLEDLCI 66
L L+K + E +YFF+I+ Y + + +I NP+FIQP S R ++ + EDL I
Sbjct 180 GLPLTK-MEEPSYFFRITNYLESIKNHIIDNPEFIQPESSRESILARLEALGSDSEDLSI 238
Query 67 SRTSFDWGIPVPIDEKHVIYVWFDALTNYLTPIGYLD---DPEK---FNKFWPADLHLVG 120
SR SF WG+PVP D++HV+YVWFDALTNY+T + + D D E+ FNK+W +H+VG
Sbjct 239 SRASFSWGVPVPNDQEHVMYVWFDALTNYITGLKWADFDNDLEENSLFNKYWENTVHIVG 298
Query 121 KEIVRFHTI 129
K+I FH++
Sbjct 299 KDITWFHSV 307
> mmu:212679 Mars2, C730026E21Rik, MetRS; methionine-tRNA synthetase
2 (mitochondrial) (EC:6.1.1.10); K01874 methionyl-tRNA
synthetase [EC:6.1.1.10]
Length=586
Score = 106 bits (265), Expect = 2e-23, Method: Composition-based stats.
Identities = 54/130 (41%), Positives = 78/130 (60%), Gaps = 7/130 (5%)
Query 2 PLSLSLSLSLSLSKEVAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGL 61
P+SL +S +KE E Y FK+S++ + L ++ NP I P ++ ++++ L
Sbjct 187 PVSLESGHPVSWTKE---ENYIFKLSQFREPLQRWLGNNPQAITPEPFHQAVLQWLEEEL 243
Query 62 EDLCISRTS--FDWGIPVPIDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLHLV 119
DL +SR S WGIPVP D+ IYVW DAL NYLT +GY D F +WPA H++
Sbjct 244 PDLSVSRRSSHLHWGIPVPGDDSQTIYVWLDALVNYLTVVGYPD--ADFKSWWPATSHII 301
Query 120 GKEIVRFHTI 129
GK+I++FH I
Sbjct 302 GKDILKFHAI 311
> hsa:92935 MARS2, MetRS, mtMetRS; methionyl-tRNA synthetase 2,
mitochondrial (EC:6.1.1.10); K01874 methionyl-tRNA synthetase
[EC:6.1.1.10]
Length=593
Score = 104 bits (259), Expect = 8e-23, Method: Composition-based stats.
Identities = 53/130 (40%), Positives = 78/130 (60%), Gaps = 7/130 (5%)
Query 2 PLSLSLSLSLSLSKEVAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGL 61
P+SL +S +KE E Y F++S++ L ++ NP I P + ++ ++ + L
Sbjct 194 PVSLESGHPVSWTKE---ENYIFRLSQFRKPLQRWLRGNPQAITPEPFHHVVLQWLDEEL 250
Query 62 EDLCISRTS--FDWGIPVPIDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLHLV 119
DL +SR S WGIPVP D+ IYVW DAL NYLT IGY + +F +WPA H++
Sbjct 251 PDLSVSRRSSHLHWGIPVPGDDSQTIYVWLDALVNYLTVIGYPN--AEFKSWWPATSHII 308
Query 120 GKEIVRFHTI 129
GK+I++FH I
Sbjct 309 GKDILKFHAI 318
> dre:566565 Methionyl-tRNA synthetase, mitochondrial-like; K01874
methionyl-tRNA synthetase [EC:6.1.1.10]
Length=595
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 47/114 (41%), Positives = 75/114 (65%), Gaps = 6/114 (5%)
Query 19 EEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQG-LEDLCISR--TSFDWGI 75
E+ Y F++S + +LL ++ +NP+ IQPV ++ ++ +++ + DL +SR + WGI
Sbjct 207 EDNYMFRLSDFRTQLLHWLRSNPNTIQPVKFQHLVLQWLEDDDIPDLSVSRQKSRLQWGI 266
Query 76 PVPIDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLHLVGKEIVRFHTI 129
PVP D + IYVW DAL NYLT +GY P+ +K+W H+VGK+I++FH I
Sbjct 267 PVPGDPEQTIYVWLDALVNYLTVVGY---PQNHSKWWSVVHHVVGKDILKFHAI 317
> xla:379528 mars2, MGC68850, metrs; methionyl-tRNA synthetase
2, mitochondrial (EC:6.1.1.10); K01874 methionyl-tRNA synthetase
[EC:6.1.1.10]
Length=562
Score = 91.3 bits (225), Expect = 7e-19, Method: Composition-based stats.
Identities = 48/115 (41%), Positives = 66/115 (57%), Gaps = 7/115 (6%)
Query 17 VAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISR--TSFDWG 74
V+EE Y F++S LL +++ P + P + +++++ L DL +SR + WG
Sbjct 194 VSEENYMFRLSSLRPALLNWLQTEP--VHPAPFLKLVHHWLEEELPDLSVSRQRSRLSWG 251
Query 75 IPVPIDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLHLVGKEIVRFHTI 129
IPVP D HVIYVW DAL NYLT GY P W HL+GK+I+RFH I
Sbjct 252 IPVPSDSSHVIYVWLDALVNYLTAAGY---PNPQLAPWGPSTHLLGKDILRFHAI 303
> sce:YGR171C MSM1; Msm1p (EC:6.1.1.10); K01874 methionyl-tRNA
synthetase [EC:6.1.1.10]
Length=575
Score = 87.8 bits (216), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 50/140 (35%), Positives = 77/140 (55%), Gaps = 28/140 (20%)
Query 18 AEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQG--LEDLCISRTS--FDW 73
+E YFF++S + +++++I NPDFI P S+R++++ ++ G L DL ISR S W
Sbjct 173 SETNYFFRLSLFNKKIVDHIRKNPDFIFPASKRDQILKELETGGTLPDLSISRPSARLKW 232
Query 74 GIPVPIDEKHVIYVWFDALTNYLTPIG----------------YLDDPEKFNKF---WPA 114
GIP P D +YVWFDAL NYL+ IG Y D + +P
Sbjct 233 GIPTPNDPSQKVYVWFDALCNYLSSIGGIPSILSNATEVVSRHYSDKSNVKGQLLIPYPK 292
Query 115 D-----LHLVGKEIVRFHTI 129
+ +H++GK+I +FHT+
Sbjct 293 EVQRNTIHVIGKDIAKFHTV 312
> bbo:BBOV_IV006630 23.m05807; methionyl-tRNA synthetase (EC:6.1.1.10);
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=522
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 50/115 (43%), Positives = 68/115 (59%), Gaps = 10/115 (8%)
Query 17 VAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWGIP 76
VAE+AYFF++SKY D LL+ I+ FI P R+NE I + L D+ I+R++ WG+P
Sbjct 184 VAEKAYFFRLSKYKDALLQLIK-REGFIVPKERQNEAIGLLSGNLPDIAITRSNTTWGVP 242
Query 77 V--PIDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLHLVGKEIVRFHTI 129
V P E H IYVW DAL +GY D ++ L L GK+I+RF T+
Sbjct 243 VGQPGFEGHTIYVWLDAL------LGYCIDRNQYG-VGTETLMLFGKDILRFMTL 290
> tpv:TP03_0137 methionyl-tRNA synthetase (EC:6.1.1.10); K01874
methionyl-tRNA synthetase [EC:6.1.1.10]
Length=691
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 43/98 (43%), Positives = 59/98 (60%), Gaps = 6/98 (6%)
Query 17 VAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWGIP 76
++E +YFF + +Y L+EYI N DFI P RNE++ +K L+DL ISRTSFDWGI
Sbjct 250 MSESSYFFTMERYRTPLVEYISTNQDFIYPKKVRNEILTRLKDQLDDLSISRTSFDWGIE 309
Query 77 VPIDEKHVIYVWFDALTNYLTPIGYLDDPEKFN--KFW 112
VP + VI + + N +GY D + FN KF+
Sbjct 310 VPSTDDIVITEYVNNDVN----LGYNHDNKLFNESKFY 343
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 33/49 (67%), Gaps = 1/49 (2%)
Query 81 EKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLHLVGKEIVRFHTI 129
EKHV+YVWFDAL+ Y+T G + + +WP DL ++GK+I F ++
Sbjct 403 EKHVMYVWFDALSFYMTSSGLGESLSEMT-YWPTDLQVIGKDISWFQSV 450
> tpv:TP01_1212 methionyl-tRNA synthetase; K01874 methionyl-tRNA
synthetase [EC:6.1.1.10]
Length=563
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/115 (35%), Positives = 66/115 (57%), Gaps = 3/115 (2%)
Query 17 VAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWGIP 76
V E AYFFK+ + +L+E+ + + + I P NE+ ++ + D+ I+R++ DWGIP
Sbjct 221 VDEPAYFFKLDIWKRKLVEFFK-DSEVILPQHSLNEVRKLLQSEINDIAITRSNCDWGIP 279
Query 77 VPIDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPA--DLHLVGKEIVRFHTI 129
+ +YVWFDAL YLT + E+ N +H++GK+I+ FHTI
Sbjct 280 ITNSGNETVYVWFDALLGYLTHLHSFPPLEQSNSNLNGLKIIHVIGKDILTFHTI 334
> pfa:PF10_0053 methionine-tRNA ligase, putative; K01874 methionyl-tRNA
synthetase [EC:6.1.1.10]
Length=749
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 54/84 (64%), Gaps = 0/84 (0%)
Query 19 EEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWGIPVP 78
E +YFF I K+ D L+++ E N +FI P R ++I +K L ++CISR + W I +P
Sbjct 301 ENSYFFNILKFKDYLIDFYEKNENFIYPPYLRKQVIYTLKNELRNICISRYNTKWAIQIP 360
Query 79 IDEKHVIYVWFDALTNYLTPIGYL 102
+ + IYVWFDAL +Y++ + YL
Sbjct 361 NEAEGTIYVWFDALLSYVSSMLYL 384
> cel:Y105E8A.20 hypothetical protein; K01874 methionyl-tRNA synthetase
[EC:6.1.1.10]
Length=524
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 40/113 (35%), Positives = 62/113 (54%), Gaps = 7/113 (6%)
Query 17 VAEEAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFD--WG 74
+ EE Y F++S++ +++ E+I + P ++ + ++ EDL ISR+ WG
Sbjct 173 IEEENYMFRLSEFREKVAEWINRENIVVHPSKYKSLALTSLEMS-EDLSISRSRSRLSWG 231
Query 75 IPVPIDEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLHLVGKEIVRFH 127
IPVP D IYVW DAL NYLT G+ + WP ++GK+I +FH
Sbjct 232 IPVPNDPTQTIYVWLDALVNYLTVTGFPNA----QSVWPPTCQVIGKDITKFH 280
> tgo:TGME49_086180 methionine tRNA synthetase, putative (EC:6.1.1.10)
Length=1761
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 21/48 (43%), Positives = 32/48 (66%), Gaps = 1/48 (2%)
Query 83 HVIYVWFDALTNYLTPIGYLD-DPEKFNKFWPADLHLVGKEIVRFHTI 129
+ +YVWFDA+ YLT G+ D + +F WP L ++GK+I+RFH +
Sbjct 1111 NAVYVWFDAVVGYLTAAGFPDVNSRRFRALWPPSLQVLGKDILRFHGV 1158
> cel:F58B3.5 mrs-1; Methionyl tRNA Synthetase family member (mrs-1);
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=917
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 35/86 (40%), Positives = 47/86 (54%), Gaps = 19/86 (22%)
Query 56 FIKQGLEDLCISRTSFDWGIPVPID--EKHVIYVWFDALTNYLTPIGYLDDP-----EKF 108
++K GL+ CI+R WG VP+D EK V YVWFDA PIGYL + +
Sbjct 258 WMKLGLDPRCITR-DLKWGTAVPLDGFEKKVFYVWFDA------PIGYLSITKCVLGDNW 310
Query 109 NKFW--PADLHL---VGKEIVRFHTI 129
K+W P ++ L VGK+ V FH +
Sbjct 311 TKWWKNPENVELFNFVGKDNVAFHAV 336
> sce:YGR264C MES1; Mes1p (EC:6.1.1.10); K01874 methionyl-tRNA
synthetase [EC:6.1.1.10]
Length=751
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 60/119 (50%), Gaps = 19/119 (15%)
Query 23 FFKISKYADRLLEYIE-ANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWGIPVPIDE 81
F + K ++ E++E A+ + + + +++K GL+ CI+R WG PVP+++
Sbjct 383 FLSLDKLESQISEWVEKASEEGNWSKNSKTITQSWLKDGLKPRCITR-DLVWGTPVPLEK 441
Query 82 --KHVIYVWFDALTNYLTPIGYLDDPEKFNKFW------PADLHL---VGKEIVRFHTI 129
V+YVWFDA IGY+ + K W P + L +GK+ V FHT+
Sbjct 442 YKDKVLYVWFDAT------IGYVSITSNYTKEWKQWWNNPEHVSLYQFMGKDNVPFHTV 494
> hsa:4141 MARS, FLJ35667, METRS, MRS, MTRNS; methionyl-tRNA synthetase
(EC:6.1.1.10); K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=900
Score = 47.8 bits (112), Expect = 8e-06, Method: Composition-based stats.
Identities = 28/82 (34%), Positives = 46/82 (56%), Gaps = 10/82 (12%)
Query 55 NFIKQGLEDLCISRTSFDWGIPVPID--EKHVIYVWFDALTNYLTPIGYLDDPEKFNKFW 112
++++ GL+ CI+R WG PVP++ E V YVWFDA YL+ D ++ ++W
Sbjct 484 SWLRDGLKPRCITR-DLKWGTPVPLEGFEDKVFYVWFDATIGYLSITANYTD--QWERWW 540
Query 113 P----ADLH-LVGKEIVRFHTI 129
DL+ + K+ V FH++
Sbjct 541 KNPEQVDLYQFMAKDNVPFHSL 562
> ath:AT4G13780 methionine--tRNA ligase, putative / methionyl-tRNA
synthetase, putative / MetRS, putative (EC:6.1.1.10); K01874
methionyl-tRNA synthetase [EC:6.1.1.10]
Length=797
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 37/119 (31%), Positives = 55/119 (46%), Gaps = 19/119 (15%)
Query 23 FFKISKYADRLLEYIEANPDFIQPVSRRNEMIN-FIKQGLEDLCISRTSFDWGIPVPIDE 81
F ++ DRL YI+ + N +++ GL CI+R WG+PVP ++
Sbjct 206 FIELPLLKDRLEAYIKKTSVTGSWSQNAIQTTNAWLRDGLRQRCITR-DLKWGVPVPHEK 264
Query 82 --KHVIYVWFDALTNYLTPIGYLDDPEKFNKFW------PADLHL---VGKEIVRFHTI 129
V YVWFDA PIGY+ + W P ++ L +GK+ V FHT+
Sbjct 265 YKDKVFYVWFDA------PIGYVSITSCYTSEWEKWWKNPENVELYQFMGKDNVPFHTV 317
> xla:379675 mars, MGC69150; methionyl-tRNA synthetase, cytoplasmic
(EC:6.1.1.10); K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=905
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 48/86 (55%), Gaps = 18/86 (20%)
Query 55 NFIKQGLEDLCISRTSFDWGIPVPID--EKHVIYVWFDALTNYLTPIGYL----DDPEKF 108
++I+ GL+ CI+R WG PVP+D V YVWFDA PIGY+ + +++
Sbjct 482 SWIRDGLKPRCITR-DLKWGTPVPLDGFRDKVFYVWFDA------PIGYISITANYTDQW 534
Query 109 NKFW--PADLHL---VGKEIVRFHTI 129
K+W P + L + K+ V FH++
Sbjct 535 EKWWKSPQQVQLYNFMAKDNVPFHSV 560
> xla:446558 mars, MGC81271, metrs, mtrns; methionyl-tRNA synthetase
(EC:6.1.1.10); K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=905
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 43/86 (50%), Gaps = 18/86 (20%)
Query 55 NFIKQGLEDLCISRTSFDWGIPVPID--EKHVIYVWFDALTNYLTPIGYLDDPEKFNKFW 112
++I+ GL+ CI+R WG PVP+D V YVWFDA PIGY+ + W
Sbjct 482 SWIRDGLKPRCITR-DLKWGTPVPLDGFRDKVFYVWFDA------PIGYISITANYTDQW 534
Query 113 ------PADLHL---VGKEIVRFHTI 129
P + L + K+ V FH++
Sbjct 535 EMWWKSPQQVQLYNFMAKDNVPFHSV 560
> mmu:216443 Mars, MGC90747, Metrs, Mtrns; methionine-tRNA synthetase
(EC:6.1.1.10); K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=902
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 28/80 (35%), Positives = 44/80 (55%), Gaps = 10/80 (12%)
Query 55 NFIKQGLEDLCISRTSFDWGIPVPID--EKHVIYVWFDALTNYLTPIGYLDDPEKFNKFW 112
++++ GL+ CI+R WG PVP++ E V YVWFDA Y++ D ++ K+W
Sbjct 486 SWLRDGLKPRCITR-DLKWGTPVPLEGFEDKVFYVWFDATIGYVSITANYTD--QWEKWW 542
Query 113 P----ADLH-LVGKEIVRFH 127
DL+ + K+ V FH
Sbjct 543 KNPEQVDLYQFMAKDNVPFH 562
> eco:b2114 metG, ECK2107, JW2101; methionyl-tRNA synthetase (EC:6.1.1.10);
K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=677
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/125 (27%), Positives = 56/125 (44%), Gaps = 24/125 (19%)
Query 20 EAYFFKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISRTSFDWGIPVPI 79
E +FF + +++ L + + Q N+M + + GL+ ISR + +G +P
Sbjct 189 EHFFFDLPSFSEMLQAWTRSGALQEQVA---NKMQEWFESGLQQWDISRDAPYFGFEIPN 245
Query 80 DEKHVIYVWFDALTNYLTPIGYL----------DDPEKFNKFWPAD-----LHLVGKEIV 124
YVW DA PIGY+ D F+++W D H +GK+IV
Sbjct 246 APGKYFYVWLDA------PIGYMGSFKNLCDKRGDSVSFDEYWKKDSTAELYHFIGKDIV 299
Query 125 RFHTI 129
FH++
Sbjct 300 YFHSL 304
> dre:338183 mars, id:ibd5106, zgc:66122; methionine-tRNA synthetase
(EC:6.1.1.10); K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=922
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 43/86 (50%), Gaps = 18/86 (20%)
Query 55 NFIKQGLEDLCISRTSFDWGIPVPIDE--KHVIYVWFDALTNYLTPIGYLDDPEKFNKFW 112
++++ GL+ CI+R WG PVP + + V YVWFDA PIGYL + W
Sbjct 482 SWLRDGLKPRCITR-DLKWGTPVPHPDYKEKVFYVWFDA------PIGYLSITANYTDQW 534
Query 113 ------PADLHL---VGKEIVRFHTI 129
P + L + K+ V FH++
Sbjct 535 ERWWKNPQQVELYNFMAKDNVPFHSV 560
> bbo:BBOV_II005760 18.m06479; tRNA synthetases class I protein
superfamily; K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1162
Score = 35.8 bits (81), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 34/77 (44%), Gaps = 17/77 (22%)
Query 44 IQPVSRRNEMINFIKQGLEDLCISRTSFDWGIPVPIDEKHVIYVWFDALTNYLTPIGYLD 103
I P +N + I +D CISR WGIP+PI +V DA LD
Sbjct 541 IYPEGAKNYLKKIIANRAQDWCISRQRI-WGIPIPI-----AFVDVDA----------LD 584
Query 104 DPEKFNKFWPADLHLVG 120
DPE K + D H+ G
Sbjct 585 DPE-LAKLYNIDYHVPG 600
> tpv:TP02_0541 hypothetical protein
Length=1221
Score = 31.2 bits (69), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 21/42 (50%), Gaps = 1/42 (2%)
Query 44 IQPVSRRNEMINFIKQGLEDLCISRTSFDWGIPVPIDEKHVI 85
I P S R + N I+ D C+SR + WG P+P + +
Sbjct 633 ITPGSARKYLNNIIRARNRDWCLSRQRY-WGAPIPYENNECL 673
> cel:K07C5.7 ttll-15; Tubulin Tyrosine Ligase Like family member
(ttll-15)
Length=513
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 44/98 (44%), Gaps = 5/98 (5%)
Query 24 FKISKYADRLLEYIEANPDFIQPVSRRNEMINFIKQGLEDLCISR-TSFDWGI---PVPI 79
F++ +LLEY E NPD + V + N N + D+ +S+ SF P+ I
Sbjct 167 FQLPAEKSKLLEYAEKNPDVLW-VQKDNTHRNIKIKSTNDMDLSKNNSFVQKFVDNPLLI 225
Query 80 DEKHVIYVWFDALTNYLTPIGYLDDPEKFNKFWPADLH 117
D K + +T+ L Y+ D + +F P D H
Sbjct 226 DNKKFDIGIYTVVTSLLPLRVYIYDGDVLIRFCPEDYH 263
> ath:AT5G08350 GRAM domain-containing protein / ABA-responsive
protein-related
Length=222
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 11/15 (73%), Positives = 12/15 (80%), Gaps = 0/15 (0%)
Query 97 TPIGYLDDPEKFNKF 111
TP+GYL DP FNKF
Sbjct 19 TPVGYLPDPASFNKF 33
> pfa:PFE0300c 60S ribosomal subunit protein L24-2, putative;
K02896 large subunit ribosomal protein L24e
Length=204
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 20/78 (25%), Positives = 34/78 (43%), Gaps = 10/78 (12%)
Query 34 LEYIEANPDFIQPVSRRN--EMINFIKQGLEDLCISRTSFDWGIPVPIDEKHVIYVWFDA 91
L YI+ NP ++ N + + KQ +D I + +F+ + H+I+ D
Sbjct 124 LNYIKKNPLLLKNTEYENVIQELTLNKQERDDTLIIKNTFE--------DDHIIFTKNDQ 175
Query 92 LTNYLTPIGYLDDPEKFN 109
Y I + D +KFN
Sbjct 176 DIKYKADISTISDKQKFN 193
> xla:378493 pak3, XPak3, pak3-A; p21 protein (Cdc42/Rac)-activated
kinase 3 (EC:2.7.11.1); K05733 p21-activated kinase 3
[EC:2.7.11.1]
Length=564
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 75 IPVPIDEKHVIYVWFDALTNYLTP 98
I +P D +H I+V FDA+T TP
Sbjct 70 ISLPSDFEHTIHVGFDAVTGEFTP 93
> ath:AT1G17230 ATP binding / protein binding / protein kinase/
protein serine/threonine kinase/ protein tyrosine kinase
Length=1101
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 21/79 (26%), Positives = 39/79 (49%), Gaps = 10/79 (12%)
Query 3 LSLSLSLSL---SLSKEVAEEAYFFKISKYAD------RLLEYIEANPDFIQPVSRRNEM 53
LS S S+S S E AY K+++ D LLE I P +QP+ + ++
Sbjct 953 LSYSKSMSAVAGSYGYIAPEYAYTMKVTEKCDIYSFGVVLLELITGKPP-VQPLEQGGDL 1011
Query 54 INFIKQGLEDLCISRTSFD 72
+N++++ + ++ + FD
Sbjct 1012 VNWVRRSIRNMIPTIEMFD 1030
Lambda K H
0.323 0.141 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044474180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40