bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7383_orf4
Length=127
Score E
Sequences producing significant alignments: (Bits) Value
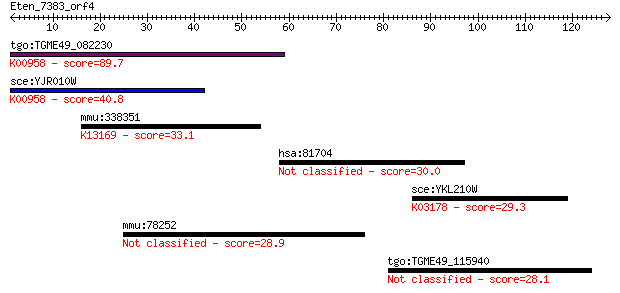
tgo:TGME49_082230 sulfate adenylyltransferas-adenylylsulfate k... 89.7 2e-18
sce:YJR010W MET3; ATP sulfurylase, catalyzes the primary step ... 40.8 0.001
mmu:338351 Akap17b, AA407619, AKAP-17B, B230333C21Rik, C230056... 33.1 0.20
hsa:81704 DOCK8, FLJ00026, FLJ00152, FLJ00346, MRD2, ZIR8; ded... 30.0 1.7
sce:YKL210W UBA1; Uba1p; K03178 ubiquitin-activating enzyme E1... 29.3 2.8
mmu:78252 Fam55b, 4432416J03Rik, AI323362; family with sequenc... 28.9 4.3
tgo:TGME49_115940 rhoptry protein, putative 28.1 6.5
> tgo:TGME49_082230 sulfate adenylyltransferas-adenylylsulfate
kinase, putative (EC:2.7.7.4 2.7.1.25); K00958 sulfate adenylyltransferase
[EC:2.7.7.4]
Length=607
Score = 89.7 bits (221), Expect = 2e-18, Method: Composition-based stats.
Identities = 37/58 (63%), Positives = 46/58 (79%), Gaps = 0/58 (0%)
Query 1 GTEFRRRLTARLPIPEWFSFPKVIEELQQFYKQNHERGLCVYFTGLPCSGAAPLHAAF 58
GT+FRR L R PIP WFSFP+V +EL++FYK N++RGLC+YFTGLPCSG + L A
Sbjct 387 GTQFRRLLEERQPIPTWFSFPEVADELRKFYKPNYQRGLCIYFTGLPCSGKSTLAQAL 444
> sce:YJR010W MET3; ATP sulfurylase, catalyzes the primary step
of intracellular sulfate activation, essential for assimilatory
reduction of sulfate to sulfide, involved in methionine
metabolism (EC:2.7.7.4); K00958 sulfate adenylyltransferase
[EC:2.7.7.4]
Length=511
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 1 GTEFRRRLTARLPIPEWFSFPKVIEELQQFYKQNHERGLCV 41
GTE RRRL IPEWFS+P+V++ L++ ++G +
Sbjct 358 GTELRRRLRVGGEIPEWFSYPEVVKILRESNPPRPKQGFSI 398
> mmu:338351 Akap17b, AA407619, AKAP-17B, B230333C21Rik, C230056F04Rik,
PRKA17B, Sfrs17b, Srsf17b, Talia; A kinase (PRKA)
anchor protein 17B; K13169 splicing factor, arginine/serine-rich
17
Length=959
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 19/44 (43%), Positives = 25/44 (56%), Gaps = 9/44 (20%)
Query 16 EWFSFPK-----VIEELQQFYKQNHERGL-CVYFTGLPCSGAAP 53
EW FPK VIE ++ Q+H++G +YF GLPC AP
Sbjct 119 EWEHFPKEKEASVIEGAEE---QDHDKGPDSIYFEGLPCKWFAP 159
> hsa:81704 DOCK8, FLJ00026, FLJ00152, FLJ00346, MRD2, ZIR8; dedicator
of cytokinesis 8
Length=1999
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 58 FRFLNKLQRSSKLLLRRYRAAALLLRRSGAAAQELLRCC 96
F+FL +RSS L RR ++ LLR + A E++ CC
Sbjct 417 FKFLADYKRSSSLQ-RRVKSIPGLLRLEISTAPEIINCC 454
> sce:YKL210W UBA1; Uba1p; K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1024
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 11/33 (33%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 86 GAAAQELLRCCCGRLSPAQGFLNSICMRVQPSP 118
G AQE+L+ C G+ +P + F+ + P P
Sbjct 368 GLVAQEVLKACSGKFTPLKQFMYFDSLESLPDP 400
> mmu:78252 Fam55b, 4432416J03Rik, AI323362; family with sequence
similarity 55, member B
Length=558
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 23/51 (45%), Gaps = 0/51 (0%)
Query 25 EELQQFYKQNHERGLCVYFTGLPCSGAAPLHAAFRFLNKLQRSSKLLLRRY 75
E Q ++HE C+ G+PC + + ++ L KLL RR+
Sbjct 239 ELCQYLDARDHEAFYCLKLPGVPCEALTHMTSKNSNISYLSLEEKLLFRRF 289
> tgo:TGME49_115940 rhoptry protein, putative
Length=999
Score = 28.1 bits (61), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 21/43 (48%), Gaps = 4/43 (9%)
Query 81 LLRRSGAAAQELLRCCCGRLSPAQGFLNSICMRVQPSPAASGW 123
+LR AA Q LLRCC +SP FL P+P S W
Sbjct 302 MLRWPRAALQLLLRCCHSLVSPGAPFLPP----TWPTPWISVW 340
Lambda K H
0.329 0.139 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2054672932
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40