bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7381_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
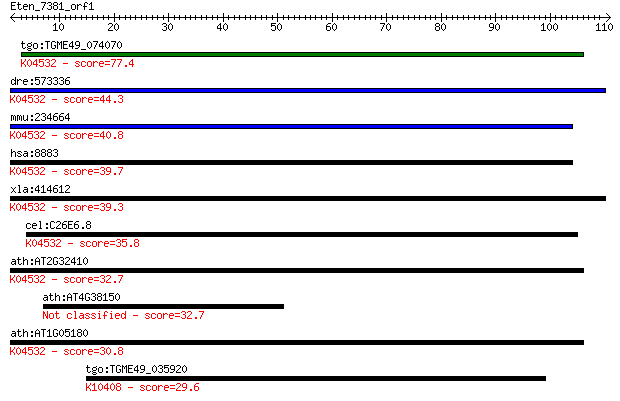
tgo:TGME49_074070 thiF family domain-containing protein ; K045... 77.4 1e-14
dre:573336 nae1, MGC171400, MGC66148, appbp1, zgc:66148; nedd8... 44.3 1e-04
mmu:234664 Nae1, 59kDa, Appbp1, MGC29435, MGC36437, MGC36630; ... 40.8 0.001
hsa:8883 NAE1, A-116A10.1, APPBP1, ula-1; NEDD8 activating enz... 39.7 0.002
xla:414612 nae1, MGC81483; NEDD8 activating enzyme E1 subunit ... 39.3 0.003
cel:C26E6.8 ula-1; yeast ULA (ubiquitin activating) homolog fa... 35.8 0.038
ath:AT2G32410 AXL; AXL (AXR1-LIKE); binding / catalytic; K0453... 32.7 0.27
ath:AT4G38150 pentatricopeptide (PPR) repeat-containing protein 32.7 0.27
ath:AT1G05180 AXR1; AXR1 (AUXIN RESISTANT 1); small protein ac... 30.8 1.0
tgo:TGME49_035920 dynein 1-beta heavy chain, flagellar inner a... 29.6 2.4
> tgo:TGME49_074070 thiF family domain-containing protein ; K04532
amyloid beta precursor protein binding protein 1
Length=779
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 48/104 (46%), Positives = 60/104 (57%), Gaps = 7/104 (6%)
Query 3 NPFYSLLQFAAAFDLEALYDKQHAHVPFVVILIQALCRYRGVTPAAAAAAAAAAAAAAAD 62
+PF L +FA +DL L D HA VPF VILIQAL RYR ++++ A A
Sbjct 314 DPFPELYEFAMEYDLNRLDDLAHAQVPFAVILIQALDRYRRTQCSSSSDACAMGEPP--- 370
Query 63 AVDLSPGP-AARQQLEKIIIEMRRSEEEENFQEALSNLHRALQP 105
L P P AR QLE II MRR +E NF EA++N+ R L+P
Sbjct 371 ---LMPLPQEARGQLEAIIQGMRRHPDEVNFDEAMANVFRILKP 411
> dre:573336 nae1, MGC171400, MGC66148, appbp1, zgc:66148; nedd8
activating enzyme E1 subunit 1; K04532 amyloid beta precursor
protein binding protein 1
Length=533
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 27/111 (24%), Positives = 53/111 (47%), Gaps = 15/111 (13%)
Query 1 ISNPFYSLLQFAAAFDLEALYDKQHAHVPFVVILIQALCRYRGVTPAAAAAAAAAAAAAA 60
+ PF L + ++DL+ + K H+H P+++++ + L ++ +
Sbjct 184 LDQPFTELKRHVESYDLDNMEKKDHSHTPWIIVVARYLEKWYNENNSQLPK--------- 234
Query 61 ADAVDLSPGPAARQQLEKIIIEMRRS--EEEENFQEALSNLHRALQPARPS 109
+ A RQ L + I++ E+EENF+EA+ N++ AL P + S
Sbjct 235 ----NYKEKEAFRQLLREGILKNENGGLEDEENFEEAIKNVNTALNPTKIS 281
> mmu:234664 Nae1, 59kDa, Appbp1, MGC29435, MGC36437, MGC36630;
NEDD8 activating enzyme E1 subunit 1; K04532 amyloid beta
precursor protein binding protein 1
Length=534
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 30/103 (29%), Positives = 50/103 (48%), Gaps = 11/103 (10%)
Query 1 ISNPFYSLLQFAAAFDLEALYDKQHAHVPFVVILIQALCRYRGVTPAAAAAAAAAAAAAA 60
+ PF L + ++DL+ + K H+H P++VI+ + L ++ T +
Sbjct 185 LDKPFPELREHLQSYDLDHMEKKDHSHTPWIVIIAKYLAQWYNET----NGRIPKSYKEK 240
Query 61 ADAVDLSPGPAARQQLEKIIIEMRRSEEEENFQEALSNLHRAL 103
D DL RQ + K E E+EENF+EA+ N++ AL
Sbjct 241 EDFRDL-----IRQGILK--NENGAPEDEENFEEAIKNVNTAL 276
> hsa:8883 NAE1, A-116A10.1, APPBP1, ula-1; NEDD8 activating enzyme
E1 subunit 1; K04532 amyloid beta precursor protein binding
protein 1
Length=528
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 30/103 (29%), Positives = 49/103 (47%), Gaps = 11/103 (10%)
Query 1 ISNPFYSLLQFAAAFDLEALYDKQHAHVPFVVILIQALCRYRGVTPAAAAAAAAAAAAAA 60
+ PF L + ++DL+ + K H+H P++VI+ + L ++ T
Sbjct 179 LDKPFPELREHFQSYDLDHMEKKDHSHTPWIVIIAKYLAQWYSET----NGRIPKTYKEK 234
Query 61 ADAVDLSPGPAARQQLEKIIIEMRRSEEEENFQEALSNLHRAL 103
D DL RQ + K E E+EENF+EA+ N++ AL
Sbjct 235 EDFRDL-----IRQGILK--NENGAPEDEENFEEAIKNVNTAL 270
> xla:414612 nae1, MGC81483; NEDD8 activating enzyme E1 subunit
1; K04532 amyloid beta precursor protein binding protein 1
Length=533
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 28/109 (25%), Positives = 52/109 (47%), Gaps = 11/109 (10%)
Query 1 ISNPFYSLLQFAAAFDLEALYDKQHAHVPFVVILIQALCRYRGVTPAAAAAAAAAAAAAA 60
+ PF L + ++DL+ + K H+H P+++++ + L ++R + +
Sbjct 184 LDQPFPELREHLQSYDLDHMERKDHSHTPWIIVVAKYLDKWR----SENGGQMPKSYKEK 239
Query 61 ADAVDLSPGPAARQQLEKIIIEMRRSEEEENFQEALSNLHRALQPARPS 109
DL RQ + K E E+EENF+EA+ N++ AL + S
Sbjct 240 ESFRDL-----IRQGILK--NENGVPEDEENFEEAIKNVNTALNITKVS 281
> cel:C26E6.8 ula-1; yeast ULA (ubiquitin activating) homolog
family member (ula-1); K04532 amyloid beta precursor protein
binding protein 1
Length=541
Score = 35.8 bits (81), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 28/107 (26%), Positives = 47/107 (43%), Gaps = 21/107 (19%)
Query 4 PFYSLLQFAAAFDLEALYDKQHAHVPFVVILIQALCRYRGVTPAAAAAAAAAAAAAAADA 63
PF L++ +L+ + +Q H P++++ +AL +R D
Sbjct 185 PFSKLIEMINETNLDEMTLEQLRHTPYILLHFKALEVFR---------------KQRNDP 229
Query 64 VDLSPGPAARQQLEKIIIEMRRSEEE------ENFQEALSNLHRALQ 104
A R++L+ I++ RRS EE ENF EA + + RA Q
Sbjct 230 EAFPSTTAERKELQAILMSFRRSSEESGTKDSENFDEAKAAVIRAFQ 276
> ath:AT2G32410 AXL; AXL (AXR1-LIKE); binding / catalytic; K04532
amyloid beta precursor protein binding protein 1
Length=417
Score = 32.7 bits (73), Expect = 0.27, Method: Composition-based stats.
Identities = 22/105 (20%), Positives = 43/105 (40%), Gaps = 17/105 (16%)
Query 1 ISNPFYSLLQFAAAFDLEALYDKQHAHVPFVVILIQALCRYRGVTPAAAAAAAAAAAAAA 60
+++P+ L + + DL H H+P+VVIL++ A A
Sbjct 180 LNSPWPELKSYVESIDLNVEEPAAHKHIPYVVILVK-----------------VAEEWAQ 222
Query 61 ADAVDLSPGPAARQQLEKIIIEMRRSEEEENFQEALSNLHRALQP 105
+ +L + + + ++ S +EEN++EAL + P
Sbjct 223 HHSGNLPSTREEKNEFKDLVKSKMVSADEENYKEALLAAFKVFAP 267
> ath:AT4G38150 pentatricopeptide (PPR) repeat-containing protein
Length=302
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 7 SLLQFAAAFDLEALYDKQHAHVPFVVILIQALCRYRGVTPAAAA 50
++L A AF E L +VP V L+ ALCR +GV A +A
Sbjct 215 NMLDDAVAFCSEMLESGHSPNVPTFVELVDALCRVKGVEQAQSA 258
> ath:AT1G05180 AXR1; AXR1 (AUXIN RESISTANT 1); small protein
activating enzyme; K04532 amyloid beta precursor protein binding
protein 1
Length=422
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 22/106 (20%), Positives = 44/106 (41%), Gaps = 18/106 (16%)
Query 1 ISNPFYSLLQFAAAFDLEALYDKQ-HAHVPFVVILIQALCRYRGVTPAAAAAAAAAAAAA 59
++NP+ L F DL H H+P+VVIL++ A A
Sbjct 92 LNNPWPELKSFVETIDLNVSEPAAAHKHIPYVVILVK-----------------MAEEWA 134
Query 60 AADAVDLSPGPAARQQLEKIIIEMRRSEEEENFQEALSNLHRALQP 105
+ + +L +++ + ++ S +E+N++EA+ + P
Sbjct 135 QSHSGNLPSTREEKKEFKDLVKSKMVSTDEDNYKEAIEAAFKVFAP 180
> tgo:TGME49_035920 dynein 1-beta heavy chain, flagellar inner
arm I1 complex, putative ; K10408 dynein heavy chain, axonemal
Length=4213
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 44/111 (39%), Gaps = 34/111 (30%)
Query 15 FDLEALYDKQHAHVPFVVILIQALCRYRGVTPAAAAAAAAAAAAAAADAVDLSPGPAARQ 74
+LEA+++ +P + +L + V P A A+A A AV L G +AR
Sbjct 3608 LNLEAVFNASKNTIPLIFVLSK-------VDPTGQLMALASAKNVQAIAVALGQGQSARA 3660
Query 75 Q---------------------------LEKIIIEMRRSEEEENFQEALSN 98
+ +EKII E+ S+ ENF+ LS+
Sbjct 3661 EKLIREGARHGFWVFLANCHLALSWLPTMEKIIEEVTESDPHENFRLWLSS 3711
Lambda K H
0.320 0.129 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40