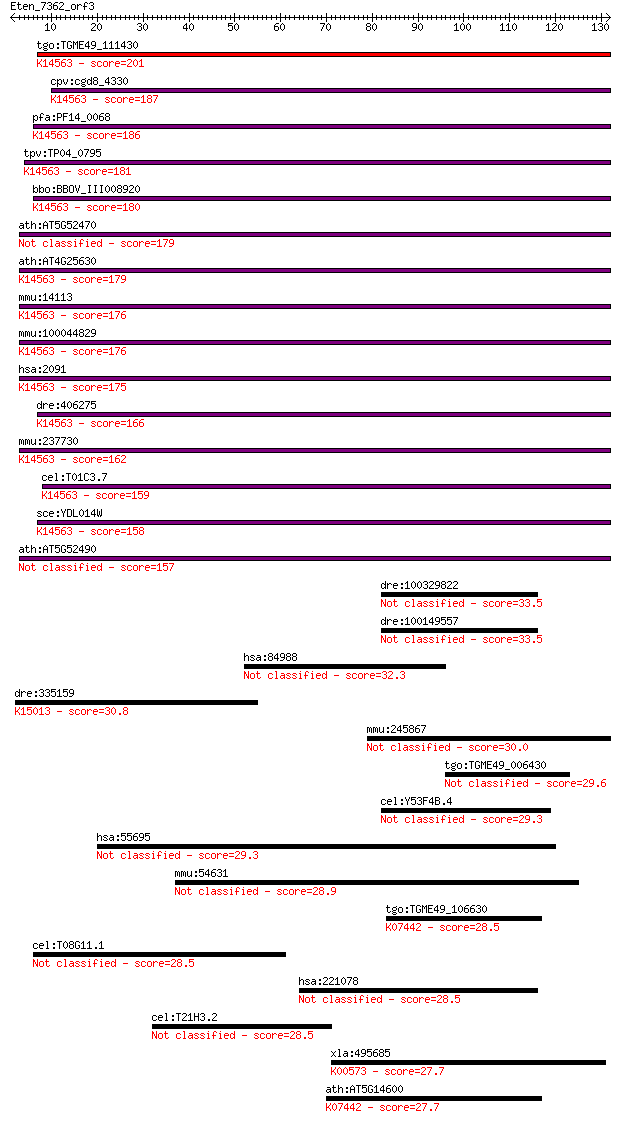
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7362_orf3
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_111430 fibrillarin, putative (EC:2.7.11.1); K14563 ... 201 4e-52
cpv:cgd8_4330 fibrillarin. RNA methylase ; K14563 rRNA 2'-O-me... 187 6e-48
pfa:PF14_0068 fibrillarin, putative; K14563 rRNA 2'-O-methyltr... 186 2e-47
tpv:TP04_0795 fibrillarin; K14563 rRNA 2'-O-methyltransferase ... 181 7e-46
bbo:BBOV_III008920 17.m07778; fibrillarin; K14563 rRNA 2'-O-me... 180 8e-46
ath:AT5G52470 FIB1; FIB1 (FIBRILLARIN 1); snoRNA binding 179 3e-45
ath:AT4G25630 FIB2; FIB2 (FIBRILLARIN 2); snoRNA binding; K145... 179 3e-45
mmu:14113 Fbl, AL022665, FIB, FLRN, RNU3IP1; fibrillarin; K145... 176 2e-44
mmu:100044829 rRNA 2'-O-methyltransferase fibrillarin-like; K1... 176 2e-44
hsa:2091 FBL, FIB, FLRN, RNU3IP1; fibrillarin; K14563 rRNA 2'-... 175 3e-44
dre:406275 fbl, wu:fb37g12, zgc:56145, zgc:77130; fibrillarin;... 166 2e-41
mmu:237730 Fbll1, AI595406, MGC62475; fibrillarin-like 1 (EC:2... 162 3e-40
cel:T01C3.7 fib-1; FIBrillarin family member (fib-1); K14563 r... 159 2e-39
sce:YDL014W NOP1, LOT3; Nop1p (EC:2.1.1.-); K14563 rRNA 2'-O-m... 158 4e-39
ath:AT5G52490 fibrillarin, putative 157 7e-39
dre:100329822 NOL1/NOP2/Sun domain family, member 6-like 33.5 0.20
dre:100149557 NOL1/NOP2/Sun domain family, member 6-like 33.5 0.20
hsa:84988 PPP1R16A, MGC14333, MYPT3; protein phosphatase 1, re... 32.3 0.39
dre:335159 acsbg2, fk81d02, im:7046047, sb:cb76, si:dkey-240a9... 30.8 1.2
mmu:245867 Pcmtd2, 5330414D10Rik, AI314201, MGC36470; protein-... 30.0 2.0
tgo:TGME49_006430 formin homology 2 domain-containing protein ... 29.6 2.6
cel:Y53F4B.4 hypothetical protein 29.3 3.2
hsa:55695 NSUN5, FLJ10267, MGC986, NOL1, NOL1R, NSUN5A, WBSCR2... 29.3 3.3
mmu:54631 Nphs1, NephrinB, nephrin; nephrosis 1 homolog, nephr... 28.9 4.2
tgo:TGME49_106630 tRNA (adenine-N(1)-)-methyltransferase catal... 28.5 5.6
cel:T08G11.1 hypothetical protein 28.5 6.2
hsa:221078 NSUN6, 4933414E04Rik, FLJ23743, NOPD1; NOP2/Sun dom... 28.5 6.2
cel:T21H3.2 ptr-16; PaTched Related family member (ptr-16) 28.5 6.3
xla:495685 pcmt1; protein-L-isoaspartate (D-aspartate) O-methy... 27.7 9.1
ath:AT5G14600 tRNA (adenine-N1-)-methyltransferase; K07442 tRN... 27.7 9.6
> tgo:TGME49_111430 fibrillarin, putative (EC:2.7.11.1); K14563
rRNA 2'-O-methyltransferase fibrillarin [EC:2.1.1.-]
Length=304
Score = 201 bits (511), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 94/125 (75%), Positives = 111/125 (88%), Gaps = 1/125 (0%)
Query 7 KAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWNPFRS 66
K V PH RFEG+F++KG+AD+L T+N+VPGESVYGEKRLE A ++ EK E+RVWNPFRS
Sbjct 68 KVLVVPH-RFEGIFMSKGKADSLVTRNMVPGESVYGEKRLEVAEESGEKIEYRVWNPFRS 126
Query 67 KLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGRDLTA 126
KLGA + GGVA+MPI+PG+KVLYLGAANGTTVSHVSDIVGPEG V+AVEFS R+GRDLT
Sbjct 127 KLGATIVGGVANMPIKPGSKVLYLGAANGTTVSHVSDIVGPEGVVYAVEFSHRSGRDLTN 186
Query 127 LAKKR 131
+AKKR
Sbjct 187 MAKKR 191
> cpv:cgd8_4330 fibrillarin. RNA methylase ; K14563 rRNA 2'-O-methyltransferase
fibrillarin [EC:2.1.1.-]
Length=301
Score = 187 bits (476), Expect = 6e-48, Method: Compositional matrix adjust.
Identities = 88/123 (71%), Positives = 107/123 (86%), Gaps = 2/123 (1%)
Query 10 VAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAA-ADAREKAEFRVWNPFRSKL 68
+ PH R EGVF+AKG++DAL T+N+VPG S+YGEKR+E + EK E+RVWNPFRSKL
Sbjct 67 IVPH-RHEGVFMAKGKSDALVTRNMVPGVSIYGEKRVETTDPETNEKIEYRVWNPFRSKL 125
Query 69 GAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGRDLTALA 128
GA + GGVA+MPI+PGAKVLYLGAANGTTVSHVSD+VGPEG+V+AVEFS R+GRDLT +A
Sbjct 126 GATIIGGVANMPIKPGAKVLYLGAANGTTVSHVSDMVGPEGSVYAVEFSHRSGRDLTDMA 185
Query 129 KKR 131
K+R
Sbjct 186 KRR 188
> pfa:PF14_0068 fibrillarin, putative; K14563 rRNA 2'-O-methyltransferase
fibrillarin [EC:2.1.1.-]
Length=318
Score = 186 bits (471), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 89/126 (70%), Positives = 104/126 (82%), Gaps = 2/126 (1%)
Query 6 GKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWNPFR 65
GK V PH RF GV++ KG++D L TKNLVPGESVYGEKR E + EK E+RVWNPFR
Sbjct 88 GKVIVVPH-RFPGVYLLKGKSDILVTKNLVPGESVYGEKRYEVMTED-EKIEYRVWNPFR 145
Query 66 SKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGRDLT 125
SKLGA L GGV +MPI+PG+KVLYLGAANGT+VSHVSD+VG EG V+AVEFS R+GRDLT
Sbjct 146 SKLGACLMGGVGNMPIKPGSKVLYLGAANGTSVSHVSDMVGDEGVVYAVEFSHRSGRDLT 205
Query 126 ALAKKR 131
++KKR
Sbjct 206 NMSKKR 211
> tpv:TP04_0795 fibrillarin; K14563 rRNA 2'-O-methyltransferase
fibrillarin [EC:2.1.1.-]
Length=287
Score = 181 bits (458), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 85/128 (66%), Positives = 104/128 (81%), Gaps = 1/128 (0%)
Query 4 GGGKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWNP 63
G K V PH RF GV++AKG++DAL T+N+V GES+YGEKR+E + EK E+R+WNP
Sbjct 47 GSNKVVVVPH-RFPGVYIAKGKSDALVTRNMVVGESIYGEKRIEVTNEDGEKIEYRMWNP 105
Query 64 FRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGRD 123
FRSKLGA + GGVA+MPI G+KVLYLGAANGTTVSHVSD+VGP G V+AVEFS R+ RD
Sbjct 106 FRSKLGATIIGGVANMPIGIGSKVLYLGAANGTTVSHVSDMVGPTGLVYAVEFSHRSARD 165
Query 124 LTALAKKR 131
LT +AK+R
Sbjct 166 LTNMAKRR 173
> bbo:BBOV_III008920 17.m07778; fibrillarin; K14563 rRNA 2'-O-methyltransferase
fibrillarin [EC:2.1.1.-]
Length=287
Score = 180 bits (457), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 83/126 (65%), Positives = 102/126 (80%), Gaps = 1/126 (0%)
Query 6 GKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWNPFR 65
K + PH RF GV++AKG++DAL T+N+VPGES+YGEKR++ + E E+RVWNPFR
Sbjct 49 NKVMIVPH-RFSGVYIAKGKSDALVTRNMVPGESIYGEKRIQITNEDSESIEYRVWNPFR 107
Query 66 SKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGRDLT 125
SKLGA + GGVA+MPI G KVLYLGAANGTTVSHVSD+VGP G V+AVEFS R+ RDLT
Sbjct 108 SKLGATIIGGVANMPIGIGTKVLYLGAANGTTVSHVSDMVGPTGMVYAVEFSHRSARDLT 167
Query 126 ALAKKR 131
+AK+R
Sbjct 168 NMAKRR 173
> ath:AT5G52470 FIB1; FIB1 (FIBRILLARIN 1); snoRNA binding
Length=273
Score = 179 bits (453), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 87/129 (67%), Positives = 103/129 (79%), Gaps = 1/129 (0%)
Query 3 RGGGKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWN 62
+GG K V PH R GVF+AKG+ DAL TKNLVPGE+VY EKR+ + K E+RVWN
Sbjct 64 KGGSKVIVEPH-RHAGVFIAKGKEDALVTKNLVPGEAVYNEKRISVQNEDGTKVEYRVWN 122
Query 63 PFRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGR 122
PFRSKL AA+ GGV ++ I+PGAKVLYLGAA+GTTVSHVSD+VGPEG V+AVEFS R+GR
Sbjct 123 PFRSKLAAAILGGVDNIWIKPGAKVLYLGAASGTTVSHVSDLVGPEGCVYAVEFSHRSGR 182
Query 123 DLTALAKKR 131
DL +AKKR
Sbjct 183 DLVNMAKKR 191
> ath:AT4G25630 FIB2; FIB2 (FIBRILLARIN 2); snoRNA binding; K14563
rRNA 2'-O-methyltransferase fibrillarin [EC:2.1.1.-]
Length=320
Score = 179 bits (453), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 87/129 (67%), Positives = 103/129 (79%), Gaps = 1/129 (0%)
Query 3 RGGGKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWN 62
+GG K V PH R GVF+AKG+ DAL TKNLVPGE+VY EKR+ + K E+RVWN
Sbjct 75 KGGSKVIVEPH-RHAGVFIAKGKEDALVTKNLVPGEAVYNEKRISVQNEDGTKTEYRVWN 133
Query 63 PFRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGR 122
PFRSKL AA+ GGV ++ I+PGAKVLYLGAA+GTTVSHVSD+VGPEG V+AVEFS R+GR
Sbjct 134 PFRSKLAAAILGGVDNIWIKPGAKVLYLGAASGTTVSHVSDLVGPEGCVYAVEFSHRSGR 193
Query 123 DLTALAKKR 131
DL +AKKR
Sbjct 194 DLVNMAKKR 202
> mmu:14113 Fbl, AL022665, FIB, FLRN, RNU3IP1; fibrillarin; K14563
rRNA 2'-O-methyltransferase fibrillarin [EC:2.1.1.-]
Length=327
Score = 176 bits (445), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 86/129 (66%), Positives = 103/129 (79%), Gaps = 2/129 (1%)
Query 3 RGGGKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWN 62
+ G V PH R EGVF+ +G+ DAL TKNLVPGESVYGEKR+ + ++ +K E+R WN
Sbjct 87 QSGKNVMVEPH-RHEGVFICRGKEDALVTKNLVPGESVYGEKRV-SISEGDDKIEYRAWN 144
Query 63 PFRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGR 122
PFRSKL AA+ GGV + I+PGAKVLYLGAA+GTTVSHVSDIVGP+G V+AVEFS R+GR
Sbjct 145 PFRSKLAAAILGGVDQIHIKPGAKVLYLGAASGTTVSHVSDIVGPDGLVYAVEFSHRSGR 204
Query 123 DLTALAKKR 131
DL LAKKR
Sbjct 205 DLINLAKKR 213
> mmu:100044829 rRNA 2'-O-methyltransferase fibrillarin-like;
K14563 rRNA 2'-O-methyltransferase fibrillarin [EC:2.1.1.-]
Length=327
Score = 176 bits (445), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 86/129 (66%), Positives = 103/129 (79%), Gaps = 2/129 (1%)
Query 3 RGGGKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWN 62
+ G V PH R EGVF+ +G+ DAL TKNLVPGESVYGEKR+ + ++ +K E+R WN
Sbjct 87 QSGKNVMVEPH-RHEGVFICRGKEDALVTKNLVPGESVYGEKRV-SISEGDDKIEYRAWN 144
Query 63 PFRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGR 122
PFRSKL AA+ GGV + I+PGAKVLYLGAA+GTTVSHVSDIVGP+G V+AVEFS R+GR
Sbjct 145 PFRSKLAAAILGGVDQIHIKPGAKVLYLGAASGTTVSHVSDIVGPDGLVYAVEFSHRSGR 204
Query 123 DLTALAKKR 131
DL LAKKR
Sbjct 205 DLINLAKKR 213
> hsa:2091 FBL, FIB, FLRN, RNU3IP1; fibrillarin; K14563 rRNA 2'-O-methyltransferase
fibrillarin [EC:2.1.1.-]
Length=321
Score = 175 bits (444), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 86/129 (66%), Positives = 103/129 (79%), Gaps = 2/129 (1%)
Query 3 RGGGKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWN 62
+ G V PH R EGVF+ +G+ DAL TKNLVPGESVYGEKR+ + ++ +K E+R WN
Sbjct 81 QSGKNVMVEPH-RHEGVFICRGKEDALVTKNLVPGESVYGEKRV-SISEGDDKIEYRAWN 138
Query 63 PFRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGR 122
PFRSKL AA+ GGV + I+PGAKVLYLGAA+GTTVSHVSDIVGP+G V+AVEFS R+GR
Sbjct 139 PFRSKLAAAILGGVDQIHIKPGAKVLYLGAASGTTVSHVSDIVGPDGLVYAVEFSHRSGR 198
Query 123 DLTALAKKR 131
DL LAKKR
Sbjct 199 DLINLAKKR 207
> dre:406275 fbl, wu:fb37g12, zgc:56145, zgc:77130; fibrillarin;
K14563 rRNA 2'-O-methyltransferase fibrillarin [EC:2.1.1.-]
Length=317
Score = 166 bits (420), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 81/125 (64%), Positives = 97/125 (77%), Gaps = 2/125 (1%)
Query 7 KAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWNPFRS 66
K V PH R EGVF+ +G+ DAL TKN+V GESVYGEKR+ + K E+R WNPFRS
Sbjct 81 KVTVEPH-RHEGVFICRGKEDALVTKNMVIGESVYGEKRINVE-EGETKIEYRAWNPFRS 138
Query 67 KLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGRDLTA 126
KL AA+ GG+ + I+PG KV+YLGAA+GTTVSHVSDIVGPEG V+AVEFS R+GRDL
Sbjct 139 KLAAAILGGIDQIHIKPGVKVMYLGAASGTTVSHVSDIVGPEGLVYAVEFSHRSGRDLLN 198
Query 127 LAKKR 131
+AKKR
Sbjct 199 VAKKR 203
> mmu:237730 Fbll1, AI595406, MGC62475; fibrillarin-like 1 (EC:2.1.1.-);
K14563 rRNA 2'-O-methyltransferase fibrillarin [EC:2.1.1.-]
Length=314
Score = 162 bits (410), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 81/129 (62%), Positives = 98/129 (75%), Gaps = 2/129 (1%)
Query 3 RGGGKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWN 62
+ G +V PH R EGVF+ +G DAL T N+VPG SVYGEKR+ + EK E+R WN
Sbjct 75 KKGITVSVEPH-RHEGVFIYRGAEDALVTLNMVPGVSVYGEKRVTVMENG-EKQEYRTWN 132
Query 63 PFRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGR 122
PFRSKL AA+ GGV + I+P +KVLYLGAA+GTTVSHVSDI+GP+G V+AVEFS RAGR
Sbjct 133 PFRSKLAAAILGGVDQIHIKPKSKVLYLGAASGTTVSHVSDIIGPDGLVYAVEFSHRAGR 192
Query 123 DLTALAKKR 131
DL +AKKR
Sbjct 193 DLVNVAKKR 201
> cel:T01C3.7 fib-1; FIBrillarin family member (fib-1); K14563
rRNA 2'-O-methyltransferase fibrillarin [EC:2.1.1.-]
Length=352
Score = 159 bits (402), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 79/124 (63%), Positives = 96/124 (77%), Gaps = 2/124 (1%)
Query 8 AAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWNPFRSK 67
V PH R GVF+ KG+ DALATKN+V GESVYGEKR+ + D E+RVWNPFRSK
Sbjct 117 VVVEPH-RLGGVFIVKGKEDALATKNMVVGESVYGEKRV-SVDDGAGSIEYRVWNPFRSK 174
Query 68 LGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGRDLTAL 127
L A++ GG+ + I+PG K+LYLGAA+GTTVSH SD+VGPEG V+AVEFS R+GRDL +
Sbjct 175 LAASIMGGLENTHIKPGTKLLYLGAASGTTVSHCSDVVGPEGIVYAVEFSHRSGRDLLGV 234
Query 128 AKKR 131
AKKR
Sbjct 235 AKKR 238
> sce:YDL014W NOP1, LOT3; Nop1p (EC:2.1.1.-); K14563 rRNA 2'-O-methyltransferase
fibrillarin [EC:2.1.1.-]
Length=327
Score = 158 bits (400), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 77/131 (58%), Positives = 97/131 (74%), Gaps = 7/131 (5%)
Query 7 KAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADARE------KAEFRV 60
K + PH R GV++A+G+ D L TKN+ PGESVYGEKR+ ++E K E+RV
Sbjct 85 KVVIEPH-RHAGVYIARGKEDLLVTKNMAPGESVYGEKRISVEEPSKEDGVPPTKVEYRV 143
Query 61 WNPFRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARA 120
WNPFRSKL A + GG+ + I PG KVLYLGAA+GT+VSHVSD+VGPEG V+AVEFS R
Sbjct 144 WNPFRSKLAAGIMGGLDELFIAPGKKVLYLGAASGTSVSHVSDVVGPEGVVYAVEFSHRP 203
Query 121 GRDLTALAKKR 131
GR+L ++AKKR
Sbjct 204 GRELISMAKKR 214
> ath:AT5G52490 fibrillarin, putative
Length=292
Score = 157 bits (397), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 77/129 (59%), Positives = 93/129 (72%), Gaps = 1/129 (0%)
Query 3 RGGGKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADAREKAEFRVWN 62
+GG K V PH R GVFVAK +ADAL TKNLVPGE +Y EKR+ + R E+RVWN
Sbjct 54 KGGSKVLVTPH-RHAGVFVAKSKADALVTKNLVPGEIIYNEKRIFVQNEDRSTVEYRVWN 112
Query 63 PFRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGR 122
P RSKL A+ GV ++ I+PG KVLYLGA++G TVSHVSDIVGPEG V+AVE S G+
Sbjct 113 PHRSKLADAITTGVDNIWIKPGVKVLYLGASSGYTVSHVSDIVGPEGCVYAVEHSDICGK 172
Query 123 DLTALAKKR 131
L +A+KR
Sbjct 173 VLMNMAEKR 181
> dre:100329822 NOL1/NOP2/Sun domain family, member 6-like
Length=417
Score = 33.5 bits (75), Expect = 0.20, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 82 RPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVE 115
RPG +VL + AA G +H++ ++G +G V A+E
Sbjct 231 RPGERVLDMCAAPGGKTTHIASLMGNQGVVVALE 264
> dre:100149557 NOL1/NOP2/Sun domain family, member 6-like
Length=417
Score = 33.5 bits (75), Expect = 0.20, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 82 RPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVE 115
RPG +VL + AA G +H++ ++G +G V A+E
Sbjct 231 RPGERVLDMCAAPGGKTTHIASLMGNQGVVVALE 264
> hsa:84988 PPP1R16A, MGC14333, MYPT3; protein phosphatase 1,
regulatory (inhibitor) subunit 16A
Length=528
Score = 32.3 bits (72), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 25/44 (56%), Gaps = 3/44 (6%)
Query 52 AREKAEFRVWNPFRSKLGAALAGGVAHMPIRPGAKVLYLGAANG 95
AR E R+ + RS+L A G H P+ GA +L++ AANG
Sbjct 203 ARAVPELRMLDDIRSRLQA---GADLHAPLDHGATLLHVAAANG 243
> dre:335159 acsbg2, fk81d02, im:7046047, sb:cb76, si:dkey-240a9.3,
wu:fj55d04, wu:fk81d02; acyl-CoA synthetase bubblegum
family member 2 (EC:6.2.1.3); K15013 long-chain-fatty-acid--CoA
ligase ACSBG [EC:6.2.1.3]
Length=752
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 24/53 (45%), Gaps = 0/53 (0%)
Query 2 PRGGGKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADARE 54
P A V HP E + + G AD+LA+ ES E E +A ARE
Sbjct 8 PTAMSTAVVMEHPGDESLLQSCGTADSLASLEDAAAESTANESSEEESACARE 60
> mmu:245867 Pcmtd2, 5330414D10Rik, AI314201, MGC36470; protein-L-isoaspartate
(D-aspartate) O-methyltransferase domain containing
2
Length=359
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 27/53 (50%), Gaps = 4/53 (7%)
Query 79 MPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGRDLTALAKKR 131
+ ++PG L LG+ G S V I+GP G VE + D+T AK++
Sbjct 76 LDLQPGLSFLNLGSGTGYLSSMVGLILGPFGVNHGVELHS----DVTEYAKQK 124
> tgo:TGME49_006430 formin homology 2 domain-containing protein
(EC:3.1.2.15 3.6.3.14)
Length=5031
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 96 TTVSHVSDIVGPEGAVFAVEFSARAGR 122
+ SH+SD G + A+FAVE RA R
Sbjct 3748 SITSHLSDSCGEDVAIFAVEACVRAAR 3774
> cel:Y53F4B.4 hypothetical protein
Length=439
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 82 RPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSA 118
RPG++V AA G SH + I+ +G V+A++ +A
Sbjct 233 RPGSQVFDTCAAPGMKTSHAAAIMENQGKVWAMDRAA 269
> hsa:55695 NSUN5, FLJ10267, MGC986, NOL1, NOL1R, NSUN5A, WBSCR20,
WBSCR20A, p120; NOP2/Sun domain family, member 5
Length=470
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 24/106 (22%), Positives = 44/106 (41%), Gaps = 8/106 (7%)
Query 20 FVAKGRADALATKNLVPGESVYGEKRLE------AAADAREKAEFRVWNPFRSKLGAALA 73
F +GRA +L + G+ + + A D E +R + + L
Sbjct 159 FSYQGRASSLDDLRALKGKHFLLDPLMPELLVFPAQTDLHEHPLYRAGHLILQDRASCLP 218
Query 74 GGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSAR 119
+ P PG+ V+ AA G SH++ ++ +G +FA + A+
Sbjct 219 AMLLDPP--PGSHVIDACAAPGNKTSHLAALLKNQGKIFAFDLDAK 262
> mmu:54631 Nphs1, NephrinB, nephrin; nephrosis 1 homolog, nephrin
(human)
Length=1256
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 39/89 (43%), Gaps = 5/89 (5%)
Query 37 GESVYGE-KRLEAAADAREKAEFRVWNPFRSKLGAALAGGVAHMPIRPGAKVLYLGAANG 95
G+S GE L A D + AE+ RS+LG L + I KVL L G
Sbjct 101 GDSAKGEFHLLIEACDLSDDAEYEC-QVGRSELGPELVSPSVILSILVSPKVLQLTPEAG 159
Query 96 TTVSHVSDIVGPEGAVFAVEFSARAGRDL 124
+TV+ V+ G E V V A+ D+
Sbjct 160 STVTWVA---GQEYVVTCVSGDAKPAPDI 185
> tgo:TGME49_106630 tRNA (adenine-N(1)-)-methyltransferase catalytic
subunit, putative (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=360
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 11/34 (32%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 83 PGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEF 116
PG ++ G +G+ H++ V P G +F EF
Sbjct 171 PGRRICEAGTGSGSLSCHLARAVAPTGHIFTFEF 204
> cel:T08G11.1 hypothetical protein
Length=3185
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 4/59 (6%)
Query 6 GKAAVAPHPRFEGVFVAKGRADALATKNLVPGESVYGEKRLEAAADARE----KAEFRV 60
G+A A H F VF+ ++D + + E + G+ + E A RE +AE RV
Sbjct 1924 GQAVDATHGEFVDVFLKNRKSDVEDRRLSIEQEEITGDLKFELAGTVRETKIGRAEKRV 1982
> hsa:221078 NSUN6, 4933414E04Rik, FLJ23743, NOPD1; NOP2/Sun domain
family, member 6
Length=469
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 64 FRSKLGAALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVE 115
F L +AL V + +PG K+L L AA G +H++ ++ +G V A++
Sbjct 217 FLQNLPSALVSHV--LNPQPGEKILDLCAAPGGKTTHIAALMHDQGEVIALD 266
> cel:T21H3.2 ptr-16; PaTched Related family member (ptr-16)
Length=881
Score = 28.5 bits (62), Expect = 6.3, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 32 KNLVPGESVYGEKRLEAAADAREKAEFRVWNPFRSKLGA 70
K L+ + V EK++EAA + R K + N RS GA
Sbjct 344 KRLMKLQKVGEEKKIEAAMERRPKQVLSIQNSIRSTAGA 382
> xla:495685 pcmt1; protein-L-isoaspartate (D-aspartate) O-methyltransferase;
K00573 protein-L-isoaspartate(D-aspartate) O-methyltransferase
[EC:2.1.1.77]
Length=228
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 27/60 (45%), Gaps = 0/60 (0%)
Query 71 ALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEFSARAGRDLTALAKK 130
A A + H + GAK L +G+ G + S +VGP+G V ++ D KK
Sbjct 66 AYALELLHDQLHEGAKALDVGSGTGILTACFSRMVGPKGKVVGIDHIKELVDDSVNNVKK 125
> ath:AT5G14600 tRNA (adenine-N1-)-methyltransferase; K07442 tRNA
(adenine-N1-)-methyltransferase catalytic subunit [EC:2.1.1.36]
Length=318
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 70 AALAGGVAHMPIRPGAKVLYLGAANGTTVSHVSDIVGPEGAVFAVEF 116
A ++ V ++ + PG VL G +G+ + ++ V P G V++ +F
Sbjct 95 ADISFVVMYLEVVPGCVVLESGTGSGSLSTSLARAVAPTGHVYSFDF 141
Lambda K H
0.317 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40