bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7355_orf1
Length=62
Score E
Sequences producing significant alignments: (Bits) Value
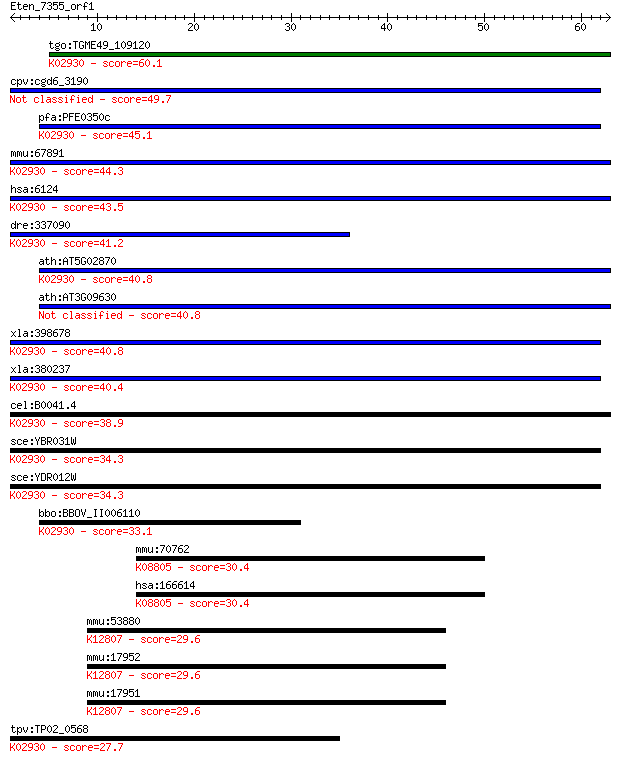
tgo:TGME49_109120 60s ribosomal protein L4, putative ; K02930 ... 60.1 2e-09
cpv:cgd6_3190 60S ribosomal protein-like 49.7 2e-06
pfa:PFE0350c 60S ribosomal protein L4, putative; K02930 large ... 45.1 6e-05
mmu:67891 Rpl4, 2010004J23Rik; ribosomal protein L4; K02930 la... 44.3 9e-05
hsa:6124 RPL4; ribosomal protein L4; K02930 large subunit ribo... 43.5 2e-04
dre:337090 rpl4, fa97a02, fl04b10, wu:fa97a02, wu:fa97c01, wu:... 41.2 0.001
ath:AT5G02870 60S ribosomal protein L4/L1 (RPL4D); K02930 larg... 40.8 0.001
ath:AT3G09630 60S ribosomal protein L4/L1 (RPL4A) 40.8 0.001
xla:398678 rpl4-b, MGC64318, l1b, rpl1b; ribosomal protein L4;... 40.8 0.001
xla:380237 rpl4-a, MGC130828, MGC53834, rpl-4, rpl1a; ribosoma... 40.4 0.001
cel:B0041.4 rpl-4; Ribosomal Protein, Large subunit family mem... 38.9 0.004
sce:YBR031W RPL4A; Rpl4ap; K02930 large subunit ribosomal prot... 34.3 0.092
sce:YDR012W RPL4B; Rpl4bp; K02930 large subunit ribosomal prot... 34.3 0.10
bbo:BBOV_II006110 18.m06507; ribosomal protein RPL4A; K02930 l... 33.1 0.22
mmu:70762 Dclk2, 6330415M09Rik, AU044875, CL2, CLICK2, Click-I... 30.4 1.4
hsa:166614 DCLK2, CL2, CLICK-II, CLICK2, CLIK2, DCAMKL2, DCDC3... 30.4 1.6
mmu:53880 Naip7, Birc1g, Naip-rs4B, Naip6; NLR family, apoptos... 29.6 2.3
mmu:17952 Naip6, Birc1f, Naip-rs4, Naip-rs4A; NLR family, apop... 29.6 2.3
mmu:17951 Naip5, Birc1e, Lgn1, Naip-rs3; NLR family, apoptosis... 29.6 2.7
tpv:TP02_0568 60S ribosomal protein L4/L1; K02930 large subuni... 27.7 9.5
> tgo:TGME49_109120 60s ribosomal protein L4, putative ; K02930
large subunit ribosomal protein L4e
Length=416
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 5 LSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPDVA 62
L++L LAPGG +GR C++TA+A ++LQ LFG +G A K+ YHL R L+ N D++
Sbjct 236 LNLLQLAPGGALGRFCLYTASAFKRLQLLFGRHTGTGTAQLKKGYHLPRALMSNADLS 293
> cpv:cgd6_3190 60S ribosomal protein-like
Length=394
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 26/61 (42%), Positives = 38/61 (62%), Gaps = 2/61 (3%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPD 60
S RL++L LAPGG GRL +FT A+++LQ+++G S + K Y L RP + N D
Sbjct 234 SVERLNLLRLAPGGTFGRLTIFTKGAIKRLQEIYG--SYTHGSSSKVGYTLPRPAMTNAD 291
Query 61 V 61
+
Sbjct 292 I 292
> pfa:PFE0350c 60S ribosomal protein L4, putative; K02930 large
subunit ribosomal protein L4e
Length=411
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 37/58 (63%), Gaps = 2/58 (3%)
Query 4 RLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPDV 61
+L++L LAPGG +GRLC+++ +A ++L ++G K+ Y L + ++ NPD+
Sbjct 234 KLNLLKLAPGGSIGRLCIWSESAFKKLDVIYGKIHEKKVT--KKNYILPKSIVHNPDI 289
> mmu:67891 Rpl4, 2010004J23Rik; ribosomal protein L4; K02930
large subunit ribosomal protein L4e
Length=419
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 40/62 (64%), Gaps = 2/62 (3%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPD 60
+ ++L++L LAPGG VGR C++T +A ++L +L+G + AA K Y+L + N D
Sbjct 231 NVSKLNILKLAPGGHVGRFCIWTESAFRKLDELYG--TWRKAASLKSNYNLPMHKMMNTD 288
Query 61 VA 62
++
Sbjct 289 LS 290
> hsa:6124 RPL4; ribosomal protein L4; K02930 large subunit ribosomal
protein L4e
Length=427
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 40/62 (64%), Gaps = 2/62 (3%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPD 60
+ ++L++L LAPGG VGR C++T +A ++L +L+G + AA K Y+L + N D
Sbjct 231 NVSKLNILKLAPGGHVGRFCIWTESAFRKLDELYG--TWRKAASLKSNYNLPMHKMINTD 288
Query 61 VA 62
++
Sbjct 289 LS 290
> dre:337090 rpl4, fa97a02, fl04b10, wu:fa97a02, wu:fa97c01, wu:fl04b10,
zgc:56659, zgc:85693; ribosomal protein L4; K02930
large subunit ribosomal protein L4e
Length=375
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLFG 35
S +RL +L LAPGG +GR C++T A ++L L+G
Sbjct 231 SVSRLDLLKLAPGGHIGRFCIWTEGAFRKLDDLYG 265
> ath:AT5G02870 60S ribosomal protein L4/L1 (RPL4D); K02930 large
subunit ribosomal protein L4e
Length=406
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 38/59 (64%), Gaps = 2/59 (3%)
Query 4 RLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPDVA 62
RL++L LAPGG +GR ++T +A ++L+ ++G S + +K+ Y L R + N D+A
Sbjct 241 RLNLLKLAPGGHLGRFVIWTKSAFEKLESIYG--SFEKPSEKKKGYVLPRAKMVNADLA 297
> ath:AT3G09630 60S ribosomal protein L4/L1 (RPL4A)
Length=405
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 38/59 (64%), Gaps = 2/59 (3%)
Query 4 RLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPDVA 62
RL++L LAPGG +GR ++T +A ++L+ ++G S + +K+ Y L R + N D+A
Sbjct 240 RLNLLKLAPGGHLGRFVIWTKSAFEKLESIYG--SFEKPSEKKKGYVLPRAKMVNADLA 296
> xla:398678 rpl4-b, MGC64318, l1b, rpl1b; ribosomal protein L4;
K02930 large subunit ribosomal protein L4e
Length=401
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 38/61 (62%), Gaps = 2/61 (3%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPD 60
+ ++L++L LAPGG VGR C++T +A ++L L+G + +A K Y+L + N D
Sbjct 235 NVSKLNLLRLAPGGHVGRFCIWTESAFRKLDDLYG--TWRKSAKLKADYNLPMHKMTNTD 292
Query 61 V 61
+
Sbjct 293 L 293
> xla:380237 rpl4-a, MGC130828, MGC53834, rpl-4, rpl1a; ribosomal
protein L4; K02930 large subunit ribosomal protein L4e
Length=396
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 38/61 (62%), Gaps = 2/61 (3%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPD 60
+ ++L++L LAPGG VGR C++T +A ++L L+G + +A K Y+L + N D
Sbjct 235 NVSKLNLLRLAPGGHVGRFCIWTESAFRKLDDLYG--TWRKSAKLKADYNLPMHKMTNTD 292
Query 61 V 61
+
Sbjct 293 L 293
> cel:B0041.4 rpl-4; Ribosomal Protein, Large subunit family member
(rpl-4); K02930 large subunit ribosomal protein L4e
Length=345
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 38/62 (61%), Gaps = 1/62 (1%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPD 60
+ RL++L LAPGG +GRL ++T +A ++L ++G ++ K+ + + P++ N D
Sbjct 230 NVERLNLLKLAPGGHLGRLIIWTESAFKKLDTIYGTTVA-NSSQLKKGWSVPLPIMANSD 288
Query 61 VA 62
+
Sbjct 289 FS 290
> sce:YBR031W RPL4A; Rpl4ap; K02930 large subunit ribosomal protein
L4e
Length=362
Score = 34.3 bits (77), Expect = 0.092, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 33/61 (54%), Gaps = 3/61 (4%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPD 60
+ A L++L LAPG +GR ++T AA +L Q++G + A K Y L ++ D
Sbjct 229 NVASLNLLQLAPGAHLGRFVIWTEAAFTKLDQVWGSET---VASSKVGYTLPSHIISTSD 285
Query 61 V 61
V
Sbjct 286 V 286
> sce:YDR012W RPL4B; Rpl4bp; K02930 large subunit ribosomal protein
L4e
Length=362
Score = 34.3 bits (77), Expect = 0.10, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 33/61 (54%), Gaps = 3/61 (4%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLFGDPSGLAAAPRKRRYHLLRPLLQNPD 60
+ A L++L LAPG +GR ++T AA +L Q++G + A K Y L ++ D
Sbjct 229 NVASLNLLQLAPGAHLGRFVIWTEAAFTKLDQVWGSET---VASSKVGYTLPSHIISTSD 285
Query 61 V 61
V
Sbjct 286 V 286
> bbo:BBOV_II006110 18.m06507; ribosomal protein RPL4A; K02930
large subunit ribosomal protein L4e
Length=360
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 4 RLSVLLLAPGGVVGRLCVFTAAALQQL 30
RL++L LAP G +GRLCV++ +A + L
Sbjct 228 RLNLLKLAPAGALGRLCVWSKSAFEAL 254
> mmu:70762 Dclk2, 6330415M09Rik, AU044875, CL2, CLICK2, Click-II,
Dcamkl2; doublecortin-like kinase 2 (EC:2.7.11.1); K08805
doublecortin and CaM kinase-like 1 [EC:2.7.11.1]
Length=771
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 18/37 (48%), Gaps = 1/37 (2%)
Query 14 GVVGRLCVFTAAALQQLQQLFGDPS-GLAAAPRKRRY 49
GVV RLC + LQ FGD +A P K RY
Sbjct 239 GVVKRLCTLDGKQVTCLQDFFGDDDVFIACGPEKYRY 275
> hsa:166614 DCLK2, CL2, CLICK-II, CLICK2, CLIK2, DCAMKL2, DCDC3,
DCDC3B, DCK2, DKFZp761I032, MGC45428; doublecortin-like
kinase 2 (EC:2.7.11.1); K08805 doublecortin and CaM kinase-like
1 [EC:2.7.11.1]
Length=766
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 18/37 (48%), Gaps = 1/37 (2%)
Query 14 GVVGRLCVFTAAALQQLQQLFGDPS-GLAAAPRKRRY 49
GVV RLC + LQ FGD +A P K RY
Sbjct 240 GVVKRLCTLDGKQVTCLQDFFGDDDVFIACGPEKFRY 276
> mmu:53880 Naip7, Birc1g, Naip-rs4B, Naip6; NLR family, apoptosis
inhibitory protein 7; K12807 baculoviral IAP repeat-containing
protein 1
Length=1402
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 25/41 (60%), Gaps = 6/41 (14%)
Query 9 LLAPGGVVGRLCVFTAAALQQLQQ----LFGDPSGLAAAPR 45
LL GG + +C+ ++++QQLQ L D SGLA+ P+
Sbjct 523 LLGAGGCISEVCL--SSSIQQLQHQVLFLLDDYSGLASLPQ 561
> mmu:17952 Naip6, Birc1f, Naip-rs4, Naip-rs4A; NLR family, apoptosis
inhibitory protein 6; K12807 baculoviral IAP repeat-containing
protein 1
Length=1403
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 25/41 (60%), Gaps = 6/41 (14%)
Query 9 LLAPGGVVGRLCVFTAAALQQLQQ----LFGDPSGLAAAPR 45
LL GG + +C+ ++++QQLQ L D SGLA+ P+
Sbjct 523 LLGAGGCISEVCL--SSSIQQLQHQVLFLLDDYSGLASLPQ 561
> mmu:17951 Naip5, Birc1e, Lgn1, Naip-rs3; NLR family, apoptosis
inhibitory protein 5; K12807 baculoviral IAP repeat-containing
protein 1
Length=1403
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 25/41 (60%), Gaps = 6/41 (14%)
Query 9 LLAPGGVVGRLCVFTAAALQQLQQ----LFGDPSGLAAAPR 45
LL GG + +C+ ++++QQLQ L D SGLA+ P+
Sbjct 523 LLGAGGCISEVCL--SSSIQQLQHQVLFLLDDYSGLASLPQ 561
> tpv:TP02_0568 60S ribosomal protein L4/L1; K02930 large subunit
ribosomal protein L4e
Length=389
Score = 27.7 bits (60), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 1 STARLSVLLLAPGGVVGRLCVFTAAALQQLQQLF 34
S L++L LAP G +GRL +++ +A + L +
Sbjct 224 SVEHLNLLKLAPAGTLGRLVLYSRSAFELLDTVL 257
Lambda K H
0.325 0.140 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2060478756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40