bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7334_orf1
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
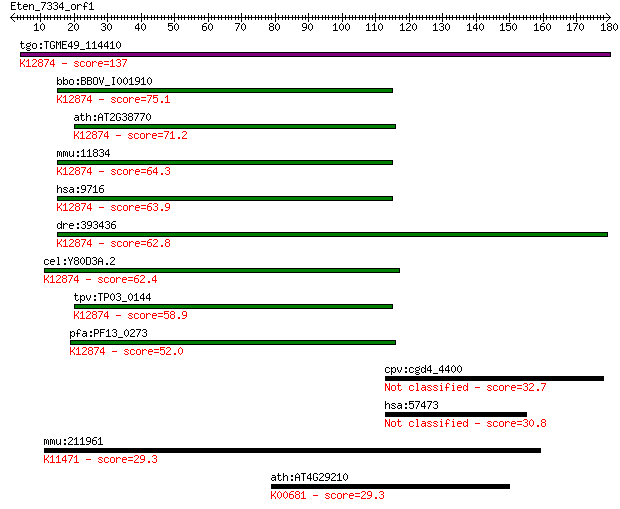
tgo:TGME49_114410 hypothetical protein ; K12874 intron-binding... 137 2e-32
bbo:BBOV_I001910 19.m02343; hypothetical protein; K12874 intro... 75.1 1e-13
ath:AT2G38770 EMB2765 (EMBRYO DEFECTIVE 2765); K12874 intron-b... 71.2 2e-12
mmu:11834 Aqr, AW495846, mKIAA0560; aquarius; K12874 intron-bi... 64.3 2e-10
hsa:9716 AQR, DKFZp686B23123, IBP160, KIAA0560, fSAP164; aquar... 63.9 3e-10
dre:393436 MGC63611; zgc:63611; K12874 intron-binding protein ... 62.8 6e-10
cel:Y80D3A.2 emb-4; abnormal EMBroygenesis family member (emb-... 62.4 8e-10
tpv:TP03_0144 hypothetical protein; K12874 intron-binding prot... 58.9 9e-09
pfa:PF13_0273 conserved Plasmodium protein, unknown function; ... 52.0 1e-06
cpv:cgd4_4400 hypothetical protein 32.7 0.70
hsa:57473 ZNF512B, GM632, KIAA1196, MGC149845, MGC149846; zinc... 30.8 2.8
mmu:211961 Asxl3, C230079D11Rik, D430002O22Rik, D930044O18Rik,... 29.3 7.0
ath:AT4G29210 GGT4; GGT4 (GAMMA-GLUTAMYL TRANSPEPTIDASE 4); ga... 29.3 8.3
> tgo:TGME49_114410 hypothetical protein ; K12874 intron-binding
protein aquarius
Length=2273
Score = 137 bits (346), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 78/176 (44%), Positives = 104/176 (59%), Gaps = 16/176 (9%)
Query 4 EDVCGSFERAEVPLQLVLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVRAERSAAAA 63
+DV + P+++V L ER+LEL +DL +QLPTRRC+LPLL + VRA +A A
Sbjct 579 DDVPLKYADEGPPMEVVYLAERMLELLIDLENQLPTRRCILPLLNDLHVVVRARMAALAK 638
Query 64 CTEAGVLRQLLQQLFFYERFEIDDDTGRPLSATACIARHYDRVEAFQRRCFELGSPAAAA 123
EAGV +Q+++ L FY RFEID+D+G PL+ C+ HY RV FQR CF L S
Sbjct 639 TPEAGVYKQMVELLDFYSRFEIDNDSGLPLTPNQCVDAHYRRVADFQRLCFSLSSDIP-- 696
Query 124 GAAAAEAATAAPNSALADAALQPISSIDDWERLQQLLQQQELLTLLDLCVHLHLLD 179
AL AL +S+ID L+ LL +QELLTLL + HL+L+D
Sbjct 697 --------------ALKLPALAAVSAIDAPAELRGLLDKQELLTLLQMATHLNLVD 738
> bbo:BBOV_I001910 19.m02343; hypothetical protein; K12874 intron-binding
protein aquarius
Length=1554
Score = 75.1 bits (183), Expect = 1e-13, Method: Composition-based stats.
Identities = 35/100 (35%), Positives = 58/100 (58%), Gaps = 0/100 (0%)
Query 15 VPLQLVLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVRAERSAAAACTEAGVLRQLL 74
+ +Q ++L E +E +DLLSQL TRR + PL+ + + VR ++ E + + +L
Sbjct 324 IAMQRLMLCENYMEFIIDLLSQLNTRRIVKPLIDAKLILVRCRQTPLHDHLEGSLFKHML 383
Query 75 QQLFFYERFEIDDDTGRPLSATACIARHYDRVEAFQRRCF 114
+ FY F ID++ G PL + R+Y+R ++FQR CF
Sbjct 384 RIFDFYMHFYIDEELGTPLDFDKVMERYYNRFDSFQRICF 423
> ath:AT2G38770 EMB2765 (EMBRYO DEFECTIVE 2765); K12874 intron-binding
protein aquarius
Length=1509
Score = 71.2 bits (173), Expect = 2e-12, Method: Composition-based stats.
Identities = 38/96 (39%), Positives = 52/96 (54%), Gaps = 0/96 (0%)
Query 20 VLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVRAERSAAAACTEAGVLRQLLQQLFF 79
VL ER +E +D+L+QLPTRR L PL+ + + SA + + QL+ L F
Sbjct 295 VLYCERFMEFLIDMLNQLPTRRYLRPLVADIAVVAKCRLSALYKHEKGKLFAQLVDLLQF 354
Query 80 YERFEIDDDTGRPLSATACIARHYDRVEAFQRRCFE 115
YE+FEI D G L+ + HYDR AFQ F+
Sbjct 355 YEKFEIKDHDGTQLTDDEALQFHYDRFMAFQLLAFK 390
> mmu:11834 Aqr, AW495846, mKIAA0560; aquarius; K12874 intron-binding
protein aquarius
Length=1481
Score = 64.3 bits (155), Expect = 2e-10, Method: Composition-based stats.
Identities = 39/101 (38%), Positives = 50/101 (49%), Gaps = 1/101 (0%)
Query 15 VPLQLVLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVRAERSAAAACTEAG-VLRQL 73
V + V ER +EL +DL + LPTRR +L L V S+ E G + QL
Sbjct 242 VTMDKVHYCERFIELMIDLEALLPTRRWFNTILDDSHLLVHCYLSSLVHREEDGHLFSQL 301
Query 74 LQQLFFYERFEIDDDTGRPLSATACIARHYDRVEAFQRRCF 114
L L FY FEI+D TG L+ HYDR+ + QR F
Sbjct 302 LDMLKFYTGFEINDQTGNALTENEMTTIHYDRITSLQRAAF 342
> hsa:9716 AQR, DKFZp686B23123, IBP160, KIAA0560, fSAP164; aquarius
homolog (mouse); K12874 intron-binding protein aquarius
Length=1485
Score = 63.9 bits (154), Expect = 3e-10, Method: Composition-based stats.
Identities = 39/101 (38%), Positives = 49/101 (48%), Gaps = 1/101 (0%)
Query 15 VPLQLVLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVRAERSAAAACTEAG-VLRQL 73
V + V ER +EL +DL + LPTRR +L L V S E G + QL
Sbjct 242 VTMDKVHYCERFIELMIDLEALLPTRRWFNTILDDSHLLVHCYLSNLVRREEDGHLFSQL 301
Query 74 LQQLFFYERFEIDDDTGRPLSATACIARHYDRVEAFQRRCF 114
L L FY FEI+D TG L+ HYDR+ + QR F
Sbjct 302 LDMLKFYTGFEINDQTGNALTENEMTTIHYDRITSLQRAAF 342
> dre:393436 MGC63611; zgc:63611; K12874 intron-binding protein
aquarius
Length=1525
Score = 62.8 bits (151), Expect = 6e-10, Method: Composition-based stats.
Identities = 50/165 (30%), Positives = 70/165 (42%), Gaps = 20/165 (12%)
Query 15 VPLQLVLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVRAERSAAAACTEAGVLR-QL 73
V ++ + ER +E +DL + LPTRR +L L V+ + S + G L QL
Sbjct 242 VSMEKIHYCERFIEFMIDLEALLPTRRWFNTVLDDAHLMVKCQLSHLTGREKEGHLFCQL 301
Query 74 LQQLFFYERFEIDDDTGRPLSATACIARHYDRVEAFQRRCFELGSPAAAAGAAAAEAATA 133
L L FY FEI D TG L+ HYDR+ + QR F
Sbjct 302 LDMLKFYTGFEISDQTGNALTQKEMTTLHYDRITSLQRAAF------------------- 342
Query 134 APNSALADAALQPISSIDDWERLQQLLQQQELLTLLDLCVHLHLL 178
A L D AL ++++D E L + TL + +L LL
Sbjct 343 AHFPELNDFALSNVAAVDTRESLNKHFGHLSPSTLHRVSAYLCLL 387
> cel:Y80D3A.2 emb-4; abnormal EMBroygenesis family member (emb-4);
K12874 intron-binding protein aquarius
Length=1467
Score = 62.4 bits (150), Expect = 8e-10, Method: Composition-based stats.
Identities = 38/108 (35%), Positives = 52/108 (48%), Gaps = 2/108 (1%)
Query 11 ERAEV--PLQLVLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVRAERSAAAACTEAG 68
E EV P+ + ER +EL +DL S L TRR +L + S+ +
Sbjct 240 EEGEVRDPIDSIKYCERFIELLIDLESILQTRRFFNSVLHSSHILTHCLLSSLISTDAGS 299
Query 69 VLRQLLQQLFFYERFEIDDDTGRPLSATACIARHYDRVEAFQRRCFEL 116
+ QL+Q L FY RFEIDD +GR L+ +HY V Q+ F L
Sbjct 300 LFFQLVQLLKFYARFEIDDLSGRQLTHKEVSEQHYQSVTRLQKAAFRL 347
> tpv:TP03_0144 hypothetical protein; K12874 intron-binding protein
aquarius
Length=1766
Score = 58.9 bits (141), Expect = 9e-09, Method: Composition-based stats.
Identities = 33/106 (31%), Positives = 54/106 (50%), Gaps = 11/106 (10%)
Query 20 VLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVR---------AERSAAAACT--EAG 68
++ +E LELFVDLLSQL RR + P+++ + L VR + AC +A
Sbjct 330 LMYIENYLELFVDLLSQLHLRRVIKPIIEYKLLLVRCKNHPLYEPVDNKTDHACNREDAN 389
Query 69 VLRQLLQQLFFYERFEIDDDTGRPLSATACIARHYDRVEAFQRRCF 114
+ +L+ L +Y F I+++ G + + +Y R FQR C+
Sbjct 390 IFIELVNILEYYLNFNINEELGTSMEYNVILEDYYKRFNKFQRVCY 435
> pfa:PF13_0273 conserved Plasmodium protein, unknown function;
K12874 intron-binding protein aquarius
Length=2533
Score = 52.0 bits (123), Expect = 1e-06, Method: Composition-based stats.
Identities = 29/97 (29%), Positives = 51/97 (52%), Gaps = 2/97 (2%)
Query 19 LVLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVRAERSAAAACTEAGVLRQLLQQLF 78
++L LE+ +E F+DLLSQL +RR L L+ + + + S + ++L++
Sbjct 652 ILLFLEKCIEFFIDLLSQLYSRRILFIFLKYFNVFIYCKISKLN--KDKRQFKELVKLFK 709
Query 79 FYERFEIDDDTGRPLSATACIARHYDRVEAFQRRCFE 115
+Y ID+ +G PL+ HY E+FQ C++
Sbjct 710 YYMEIYIDNFSGNPLTYLEINNEHYKNFESFQIFCYK 746
> cpv:cgd4_4400 hypothetical protein
Length=286
Score = 32.7 bits (73), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 27/65 (41%), Gaps = 1/65 (1%)
Query 113 CFELGSPAAAAGAAAAEAATAAPNSALADAALQPISSIDDWERLQQLLQQQELLTLLDLC 172
CFE G ++ T + NS +D +S W R+ Q+L + +LL+
Sbjct 186 CFE-NCTEDIGGLISSHQKTGSGNSPRSDIVQSSVSKSGVWNRISQVLPRDSYFSLLNFV 244
Query 173 VHLHL 177
HL
Sbjct 245 RQYHL 249
> hsa:57473 ZNF512B, GM632, KIAA1196, MGC149845, MGC149846; zinc
finger protein 512B
Length=892
Score = 30.8 bits (68), Expect = 2.8, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Query 113 CFELGSPAAAAGAAAAEAATAAPNSALADAALQPISSIDDWE 154
CF+ GSPA+ + A N AL D P+S ++DW+
Sbjct 58 CFDPGSPASDKTEGKKKGRPKAENQALRDI---PLSLMNDWK 96
> mmu:211961 Asxl3, C230079D11Rik, D430002O22Rik, D930044O18Rik,
Gm329, Gm945; additional sex combs like 3 (Drosophila); K11471
additional sex combs-like protein
Length=2204
Score = 29.3 bits (64), Expect = 7.0, Method: Composition-based stats.
Identities = 34/148 (22%), Positives = 59/148 (39%), Gaps = 4/148 (2%)
Query 11 ERAEVPLQLVLLLERLLELFVDLLSQLPTRRCLLPLLQQQQLAVRAERSAAAACTEAGVL 70
E+ E P + + ++ LE+ D L P+ + ++Q++ E SA A L
Sbjct 466 EKLECPQEEMSVVIDQLEI-CDSLLPCPSSVTHILDVEQKEQETTIETSAMALREGPSSL 524
Query 71 RQLLQQLFFYERFEIDDDTGRPLSATACIARHYDRVEAFQRRCFELGSPAAAAGAAAAEA 130
L E+ D LS ACI+ E+ + C L SP + + E+
Sbjct 525 ESQLPNEGIAVDMELQSDPEEQLSENACISETSFSSESPEGACASLPSPGGETQSTSEES 584
Query 131 ATAAPNSALADAALQPISSIDDWERLQQ 158
T A +L +SS ++ ++ Q
Sbjct 585 CTPA---SLETTFCSEVSSTENTDKYNQ 609
> ath:AT4G29210 GGT4; GGT4 (GAMMA-GLUTAMYL TRANSPEPTIDASE 4);
gamma-glutamyltransferase/ glutathione gamma-glutamylcysteinyltransferase
(EC:2.3.2.2); K00681 gamma-glutamyltranspeptidase
[EC:2.3.2.2]
Length=637
Score = 29.3 bits (64), Expect = 8.3, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 31/72 (43%), Gaps = 4/72 (5%)
Query 79 FYERFEIDDDTGRPLSATACIARHYDRV-EAFQRRCFELGSPAAAAGAAAAEAATAAPNS 137
F R IDDD L + + + V A RC E+G+ +G A +AA A
Sbjct 55 FLSRQAIDDDHSLSLGTISDVVESENGVVAADDARCSEIGASVLRSGGHAVDAAVAI--- 111
Query 138 ALADAALQPISS 149
L + P+SS
Sbjct 112 TLCVGVVNPMSS 123
Lambda K H
0.323 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4795148792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40