bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7327_orf1
Length=111
Score E
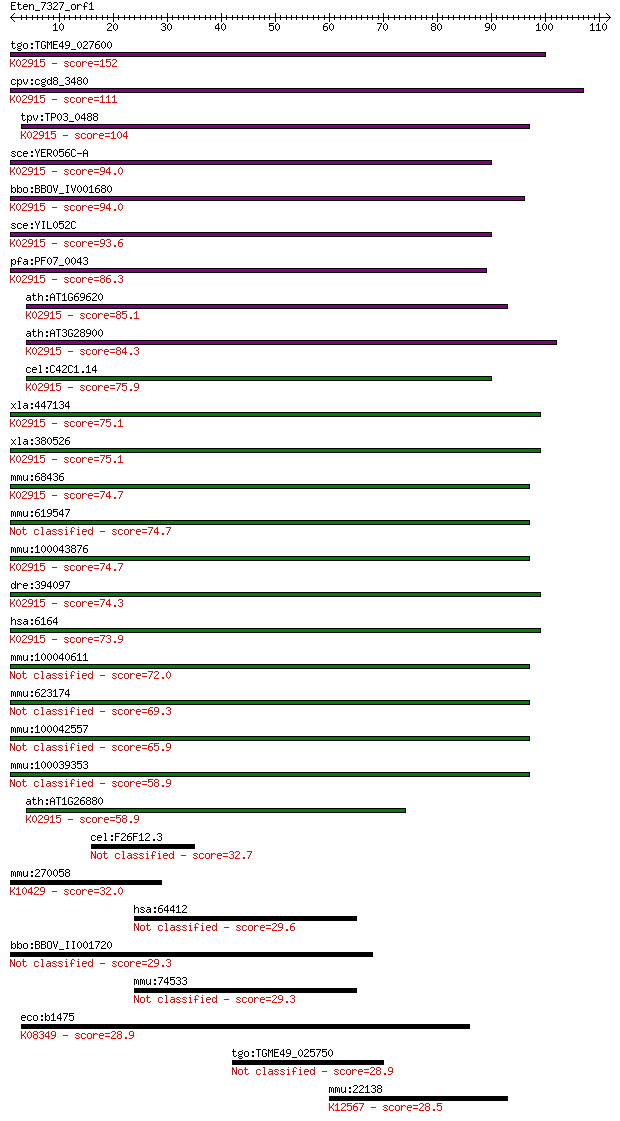
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_027600 ribosomal protein L34, putative ; K02915 lar... 152 3e-37
cpv:cgd8_3480 60S ribosomal protein L34 ; K02915 large subunit... 111 5e-25
tpv:TP03_0488 60S ribosomal protein L34a; K02915 large subunit... 104 8e-23
sce:YER056C-A RPL34A; L34A; K02915 large subunit ribosomal pro... 94.0 1e-19
bbo:BBOV_IV001680 21.m02928; ribosomal protein L34; K02915 lar... 94.0 1e-19
sce:YIL052C RPL34B; L34B; K02915 large subunit ribosomal prote... 93.6 1e-19
pfa:PF07_0043 60S ribosomal protein L34-A, putative; K02915 la... 86.3 2e-17
ath:AT1G69620 RPL34; RPL34 (RIBOSOMAL PROTEIN L34); structural... 85.1 5e-17
ath:AT3G28900 60S ribosomal protein L34 (RPL34C); K02915 large... 84.3 8e-17
cel:C42C1.14 rpl-34; Ribosomal Protein, Large subunit family m... 75.9 3e-14
xla:447134 rpl34, MGC85388; ribosomal protein L34; K02915 larg... 75.1 6e-14
xla:380526 rpl34, MGC64481, xl34; ribosomal protein L34; K0291... 75.1 6e-14
mmu:68436 Rpl34, 1100001I22Rik, MGC103270; ribosomal protein L... 74.7 7e-14
mmu:619547 Rpl34-ps1, EG619547, Gm10239, MGC103270, MGC117759;... 74.7 7e-14
mmu:100043876 Gm4705; predicted gene 4705; K02915 large subuni... 74.7 7e-14
dre:394097 rpl34, MGC56455, zgc:56455, zgc:77706; ribosomal pr... 74.3 9e-14
hsa:6164 RPL34, MGC111005; ribosomal protein L34; K02915 large... 73.9 1e-13
mmu:100040611 Gm10154; predicted gene 10154 72.0
mmu:623174 Gm6404, EG623174; predicted gene 6404 69.3
mmu:100042557 Gm10086; ribosomal protein L34 pseudogene 65.9 3e-11
mmu:100039353 Gm2178; predicted gene 2178 58.9
ath:AT1G26880 60S ribosomal protein L34 (RPL34A); K02915 large... 58.9 4e-09
cel:F26F12.3 hypothetical protein 32.7 0.31
mmu:270058 Mtap1s, 6430517J16Rik, Bpy2ip1, MAP1S, Map8, VCY2IP... 32.0 0.49
hsa:64412 GZF1, ZBTB23, ZNF336; GDNF-inducible zinc finger pro... 29.6 2.5
bbo:BBOV_II001720 18.m06132; serine threonine phosphatase 2C c... 29.3 3.3
mmu:74533 Gzf1, 8430437G08Rik, Zfp336, mGZF1; GDNF-inducible z... 29.3 3.6
eco:b1475 fdnH, ECK1469, JW1471; formate dehydrogenase-N, Fe-S... 28.9 4.1
tgo:TGME49_025750 hypothetical protein 28.9 4.4
mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik, 23... 28.5 6.2
> tgo:TGME49_027600 ribosomal protein L34, putative ; K02915 large
subunit ribosomal protein L34e
Length=134
Score = 152 bits (384), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 74/99 (74%), Positives = 88/99 (88%), Gaps = 0/99 (0%)
Query 1 DRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYG 60
+RVRRV+TPGAKVV+ NV KQ SR +CG C R LPGIPA P + RLLKKRERTV+RAYG
Sbjct 18 NRVRRVRTPGAKVVFHNVGKQPSRPKCGNCHRALPGIPAVAPHRLRLLKKRERTVHRAYG 77
Query 61 GTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKRDEV 99
G+RCH+CVRE+++RAFLVEEQKCVKQVLQVKE+QK+DE+
Sbjct 78 GSRCHACVRERIVRAFLVEEQKCVKQVLQVKEKQKKDEM 116
> cpv:cgd8_3480 60S ribosomal protein L34 ; K02915 large subunit
ribosomal protein L34e
Length=138
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 61/107 (57%), Positives = 79/107 (73%), Gaps = 1/107 (0%)
Query 1 DRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYG 60
+RVR VKTPG + + N+ K SR +CG C + L GIPA P + + LKKRERTV RAYG
Sbjct 21 NRVRVVKTPGGRNLIQNIGKVYSRPKCGDCKKPLAGIPACAPYEMKHLKKRERTVARAYG 80
Query 61 GTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKRDEVKTE-NKKS 106
GT+C +CVR+++LRAFL+EEQK VK V+ KE+QK+ E K + NKKS
Sbjct 81 GTKCPTCVRQRILRAFLLEEQKVVKSVIAEKEQQKKSEEKPKVNKKS 127
> tpv:TP03_0488 60S ribosomal protein L34a; K02915 large subunit
ribosomal protein L34e
Length=138
Score = 104 bits (259), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 52/94 (55%), Positives = 73/94 (77%), Gaps = 2/94 (2%)
Query 3 VRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGT 62
V+RV+TPGA++V N++K R RCG C R L G+ A RP ++ LK+R RTV+R YGG+
Sbjct 20 VKRVRTPGARLVLHNLKKVGKRPRCGDCKRTLAGVSAVRPFLYKNLKRRNRTVSRPYGGS 79
Query 63 RCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKR 96
RCH+CV+++++RAFL EEQKCV++V VKER+ R
Sbjct 80 RCHNCVKDRIIRAFLKEEQKCVRRV--VKERETR 111
> sce:YER056C-A RPL34A; L34A; K02915 large subunit ribosomal protein
L34e
Length=121
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 66/89 (74%), Gaps = 0/89 (0%)
Query 1 DRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYG 60
++++ VKTPG + +V+K A+R +CG CG L GI RP Q+ + K +TV+RAYG
Sbjct 18 NKIKVVKTPGGILRAQHVKKLATRPKCGDCGSALQGISTLRPRQYATVSKTHKTVSRAYG 77
Query 61 GTRCHSCVREKVLRAFLVEEQKCVKQVLQ 89
G+RC +CV+E+++RAFL+EEQK VK+V++
Sbjct 78 GSRCANCVKERIIRAFLIEEQKIVKKVVK 106
> bbo:BBOV_IV001680 21.m02928; ribosomal protein L34; K02915 large
subunit ribosomal protein L34e
Length=157
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 49/95 (51%), Positives = 68/95 (71%), Gaps = 0/95 (0%)
Query 1 DRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYG 60
++ R+VKTPG ++V K A +CG C L GIPA RP +R LKKRER V+RAYG
Sbjct 18 NKQRKVKTPGGRLVVQRRVKVAQGPKCGDCKCKLAGIPALRPHLYRNLKKRERRVSRAYG 77
Query 61 GTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQK 95
G RCH CV+++++RAFL EEQK +K+V++ +E +K
Sbjct 78 GVRCHKCVKDRIIRAFLKEEQKALKKVIEEREAKK 112
> sce:YIL052C RPL34B; L34B; K02915 large subunit ribosomal protein
L34e
Length=121
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 43/89 (48%), Positives = 66/89 (74%), Gaps = 0/89 (0%)
Query 1 DRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYG 60
++++ VKTPG + +V+K A+R +CG CG L GI RP Q+ + K +TV+RAYG
Sbjct 18 NKIKVVKTPGGILRAQHVKKLATRPKCGDCGSALQGISTLRPRQYATVSKTHKTVSRAYG 77
Query 61 GTRCHSCVREKVLRAFLVEEQKCVKQVLQ 89
G+RC +CV+E+++RAFL+EEQK VK+V++
Sbjct 78 GSRCANCVKERIVRAFLIEEQKIVKKVVK 106
> pfa:PF07_0043 60S ribosomal protein L34-A, putative; K02915
large subunit ribosomal protein L34e
Length=150
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 41/88 (46%), Positives = 60/88 (68%), Gaps = 0/88 (0%)
Query 1 DRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYG 60
++VR V+TPG K+ V+K+A + +C C + G+ A RP +++ RTV RAYG
Sbjct 18 NKVRPVRTPGGKLTIHVVKKKAGKPKCADCKTAIQGVKALRPADNYRARRKNRTVARAYG 77
Query 61 GTRCHSCVREKVLRAFLVEEQKCVKQVL 88
G+ C C+RE+++RAFL EEQKCV+QVL
Sbjct 78 GSICARCIRERIMRAFLFEEQKCVRQVL 105
> ath:AT1G69620 RPL34; RPL34 (RIBOSOMAL PROTEIN L34); structural
constituent of ribosome; K02915 large subunit ribosomal protein
L34e
Length=119
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 45/91 (49%), Positives = 63/91 (69%), Gaps = 2/91 (2%)
Query 4 RRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRL--LKKRERTVNRAYGG 61
R VKTPG K+ Y +K+AS +C G+ + GIP RP +++ L + RTVNRAYGG
Sbjct 21 RIVKTPGGKLTYQTTKKRASGPKCPVTGKRIQGIPHLRPTEYKRSRLSRNRRTVNRAYGG 80
Query 62 TRCHSCVREKVLRAFLVEEQKCVKQVLQVKE 92
S VRE+++RAFLVEEQK VK+VL++++
Sbjct 81 VLSGSAVRERIIRAFLVEEQKIVKKVLKLQK 111
> ath:AT3G28900 60S ribosomal protein L34 (RPL34C); K02915 large
subunit ribosomal protein L34e
Length=120
Score = 84.3 bits (207), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 46/100 (46%), Positives = 67/100 (67%), Gaps = 2/100 (2%)
Query 4 RRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRL--LKKRERTVNRAYGG 61
R VKTPG K+ Y K+AS +C G+ + GIP RP +++ L + ERTVNRAYGG
Sbjct 21 RIVKTPGGKLTYQTTNKRASGPKCPVTGKRIQGIPHLRPAEYKRSRLARNERTVNRAYGG 80
Query 62 TRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKRDEVKT 101
VRE+++RAFLVEEQK VK+VL++++ +++ K+
Sbjct 81 VLSGVAVRERIVRAFLVEEQKIVKKVLKLQKAKEKTAPKS 120
> cel:C42C1.14 rpl-34; Ribosomal Protein, Large subunit family
member (rpl-34); K02915 large subunit ribosomal protein L34e
Length=110
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 41/86 (47%), Positives = 55/86 (63%), Gaps = 0/86 (0%)
Query 4 RRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTR 63
R VKTPG ++V ++K+ +C G L GI RP RLLK+ ERTV RAYGG
Sbjct 21 RLVKTPGGRLVVQYIKKRGQIPKCRDTGVKLHGITPARPIALRLLKRNERTVTRAYGGCL 80
Query 64 CHSCVREKVLRAFLVEEQKCVKQVLQ 89
+ V+E++ RAFLVEEQK V +V++
Sbjct 81 SPNAVKERITRAFLVEEQKIVNKVIK 106
> xla:447134 rpl34, MGC85388; ribosomal protein L34; K02915 large
subunit ribosomal protein L34e
Length=117
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 41/100 (41%), Positives = 64/100 (64%), Gaps = 2/100 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +TPG ++VYL +K +A + CG C L GI A RP L K ++ V+RA
Sbjct 18 NKTRLSRTPGNRIVYLYTKKVGKAPKSACGICPGRLRGIRAVRPKVLMRLSKTKKHVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKRDE 98
YGG+ C CVR+++ RAFL+EEQK V +VL+ + + ++ +
Sbjct 78 YGGSMCAKCVRDRIKRAFLIEEQKIVVKVLKAQAQSQKAK 117
> xla:380526 rpl34, MGC64481, xl34; ribosomal protein L34; K02915
large subunit ribosomal protein L34e
Length=117
Score = 75.1 bits (183), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 41/100 (41%), Positives = 64/100 (64%), Gaps = 2/100 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +TPG ++VYL +K +A + CG C L GI A RP L K ++ V+RA
Sbjct 18 NKTRLSRTPGNRIVYLYTKKVGKAPKSACGICPGRLRGIRAVRPKVLMRLSKTKKHVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKRDE 98
YGG+ C CVR+++ RAFL+EEQK V +VL+ + + ++ +
Sbjct 78 YGGSMCAKCVRDRIKRAFLIEEQKIVVKVLKAQAQSQKAK 117
> mmu:68436 Rpl34, 1100001I22Rik, MGC103270; ribosomal protein
L34; K02915 large subunit ribosomal protein L34e
Length=117
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 40/98 (40%), Positives = 63/98 (64%), Gaps = 2/98 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +TPG ++VYL +K +A + CG C L G+ A RP L K ++ V+RA
Sbjct 18 NKTRLSRTPGNRIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKR 96
YGG+ C CVR+++ RAFL+EEQK V +VL+ + + ++
Sbjct 78 YGGSMCAKCVRDRIKRAFLIEEQKIVVKVLKAQAQSQK 115
> mmu:619547 Rpl34-ps1, EG619547, Gm10239, MGC103270, MGC117759;
ribosomal protein L34, pseudogene 1
Length=117
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 40/98 (40%), Positives = 63/98 (64%), Gaps = 2/98 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +TPG ++VYL +K +A + CG C L G+ A RP L K ++ V+RA
Sbjct 18 NKTRLSRTPGNRIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKR 96
YGG+ C CVR+++ RAFL+EEQK V +VL+ + + ++
Sbjct 78 YGGSMCAKCVRDRIKRAFLIEEQKIVVKVLKAQAQSQK 115
> mmu:100043876 Gm4705; predicted gene 4705; K02915 large subunit
ribosomal protein L34e
Length=117
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 40/98 (40%), Positives = 63/98 (64%), Gaps = 2/98 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +TPG ++VYL +K +A + CG C L G+ A RP L K ++ V+RA
Sbjct 18 NKTRLSRTPGNRIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKR 96
YGG+ C CVR+++ RAFL+EEQK V +VL+ + + ++
Sbjct 78 YGGSMCAKCVRDRIKRAFLIEEQKIVVKVLKAQAQSQK 115
> dre:394097 rpl34, MGC56455, zgc:56455, zgc:77706; ribosomal
protein L34; K02915 large subunit ribosomal protein L34e
Length=117
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 40/100 (40%), Positives = 64/100 (64%), Gaps = 2/100 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +TPG ++VYL +K ++ + CG C L GI A RP L K ++ V+RA
Sbjct 18 NKTRLSRTPGNRIVYLYTKKTGKSPKSACGICPGRLRGIRAVRPQVLMRLSKTKKHVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKRDE 98
YGG+ C CVR+++ RAFL+EEQK V +VL+ + + ++ +
Sbjct 78 YGGSMCAKCVRDRIKRAFLIEEQKIVVKVLKAQAQTQKSK 117
> hsa:6164 RPL34, MGC111005; ribosomal protein L34; K02915 large
subunit ribosomal protein L34e
Length=117
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 40/100 (40%), Positives = 64/100 (64%), Gaps = 2/100 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +TPG ++VYL +K +A + CG C L G+ A RP L K ++ V+RA
Sbjct 18 NKTRLSRTPGNRIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTKKHVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKRDE 98
YGG+ C CVR+++ RAFL+EEQK V +VL+ + + ++ +
Sbjct 78 YGGSMCAKCVRDRIKRAFLIEEQKIVVKVLKAQAQSQKAK 117
> mmu:100040611 Gm10154; predicted gene 10154
Length=117
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 39/98 (39%), Positives = 62/98 (63%), Gaps = 2/98 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +TPG ++VYL +K +A + CG C L G+ A RP L K ++ V+RA
Sbjct 18 NKTRLSRTPGNRIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKR 96
Y G+ C CVR+++ RAFL+EEQK V +VL+ + + ++
Sbjct 78 YDGSMCAKCVRDRIKRAFLIEEQKIVVKVLKAQAQSQK 115
> mmu:623174 Gm6404, EG623174; predicted gene 6404
Length=117
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 61/98 (62%), Gaps = 2/98 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +TPG ++VYL +K +A + CG C L G+ A RP L K ++ V+RA
Sbjct 18 NKTRLSRTPGNRIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKQVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKR 96
YG + C C+R+++ AFL+EEQK V +VL+ + + ++
Sbjct 78 YGSSMCAKCIRDRIKGAFLIEEQKIVVKVLKAQAQSQK 115
> mmu:100042557 Gm10086; ribosomal protein L34 pseudogene
Length=117
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 61/98 (62%), Gaps = 2/98 (2%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRA 58
++ R +T G ++VYL +K +A + CG C L G+ A RP L K ++ V+RA
Sbjct 18 NKTRLSRTLGNRIVYLYTKKVGKAPKSACGVCPGRLRGVRAVRPKVLMRLSKTQKHVSRA 77
Query 59 YGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKR 96
YGG+ C CV +++ +AFL+EEQK V +VL+ + + ++
Sbjct 78 YGGSMCAKCVCDRIKQAFLIEEQKIVVKVLKAQAQSQK 115
> mmu:100039353 Gm2178; predicted gene 2178
Length=118
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 34/99 (34%), Positives = 58/99 (58%), Gaps = 3/99 (3%)
Query 1 DRVRRVKTPGAKVVYLNVRK--QASRQRCGGCGRLLPGIPARRPPQFRL-LKKRERTVNR 57
++ R +TP ++VYL +K +A + CG C L G+PA R + L K + V+R
Sbjct 18 NKTRLSRTPDNRIVYLYTKKVGKAPKSACGVCPLRLQGLPAVRARVVLMRLSKTKMQVSR 77
Query 58 AYGGTRCHSCVREKVLRAFLVEEQKCVKQVLQVKERQKR 96
AY + C CV +++ +AF +EEQK + VL+V+ + ++
Sbjct 78 AYSCSMCAKCVCDRIKQAFFIEEQKIIVNVLKVQAQSQK 116
> ath:AT1G26880 60S ribosomal protein L34 (RPL34A); K02915 large
subunit ribosomal protein L34e
Length=96
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 45/72 (62%), Gaps = 2/72 (2%)
Query 4 RRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFR--LLKKRERTVNRAYGG 61
R VKTPG K+VY +K+AS +C G+ + GIP RP +++ L + RTVNRAYGG
Sbjct 21 RIVKTPGGKLVYQTTKKRASGPKCPVTGKRIQGIPHLRPSEYKRSRLSRNRRTVNRAYGG 80
Query 62 TRCHSCVREKVL 73
S VRE+ +
Sbjct 81 VLSGSAVRERFV 92
> cel:F26F12.3 hypothetical protein
Length=890
Score = 32.7 bits (73), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 12/19 (63%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 16 LNVRKQASRQRCGGCGRLL 34
+ VR QASR CGGCGR+
Sbjct 708 MEVRDQASRAECGGCGRVF 726
> mmu:270058 Mtap1s, 6430517J16Rik, Bpy2ip1, MAP1S, Map8, VCY2IP1;
microtubule-associated protein 1S; K10429 microtubule-associated
protein 1
Length=973
Score = 32.0 bits (71), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 1 DRVRRVKTPGAKVVYLNVRKQASRQRCG 28
+R+RR+ +P VV+LN R+ ASR R G
Sbjct 317 ERLRRLLSPALGVVFLNAREAASRLRGG 344
> hsa:64412 GZF1, ZBTB23, ZNF336; GDNF-inducible zinc finger protein
1
Length=711
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 20/41 (48%), Gaps = 6/41 (14%)
Query 24 RQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRC 64
R RCG CG+ L A R L +R T +R YG T C
Sbjct 406 RHRCGQCGKGLSSKTALR------LHERTHTGDRPYGCTEC 440
> bbo:BBOV_II001720 18.m06132; serine threonine phosphatase 2C
containing protein
Length=339
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 26/67 (38%), Gaps = 10/67 (14%)
Query 1 DRVRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYG 60
D RR+K G VVY R + RCG L RP Q + +RA+G
Sbjct 190 DEKRRIKAAGGIVVYSKTRHATTFLRCG--DYLKRHARGERPMQIQY--------SRAFG 239
Query 61 GTRCHSC 67
G C
Sbjct 240 GKELKRC 246
> mmu:74533 Gzf1, 8430437G08Rik, Zfp336, mGZF1; GDNF-inducible
zinc finger protein 1
Length=706
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 21/41 (51%), Gaps = 6/41 (14%)
Query 24 RQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGTRC 64
R RCG CG+ L A R L +R T ++ YG T+C
Sbjct 404 RHRCGQCGKGLSSKTALR------LHERTHTGDKPYGCTKC 438
> eco:b1475 fdnH, ECK1469, JW1471; formate dehydrogenase-N, Fe-S
(beta) subunit, nitrate-inducible; K08349 formate dehydrogenase-N,
beta subunit [EC:1.2.1.2]
Length=294
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 34/83 (40%), Gaps = 14/83 (16%)
Query 3 VRRVKTPGAKVVYLNVRKQASRQRCGGCGRLLPGIPARRPPQFRLLKKRERTVNRAYGGT 62
++ + GA + Y N + C GCG + G P P RL K+ NR Y T
Sbjct 109 LKACPSAGAIIQYANGIVDFQSENCIGCGYCIAGCPFNIP---RLNKED----NRVYKCT 161
Query 63 RCHSCVREKVLRAFLVEEQKCVK 85
C V R + +E CVK
Sbjct 162 LC-------VDRVSVGQEPACVK 177
> tgo:TGME49_025750 hypothetical protein
Length=7371
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 18/30 (60%), Gaps = 2/30 (6%)
Query 42 PPQFRLLKKRERTVNRAYG--GTRCHSCVR 69
PP+ R+ R+R + + G TRCHS VR
Sbjct 5067 PPRLRVASNRQRCASNSRGDRSTRCHSLVR 5096
> mmu:22138 Ttn, 1100001C23Rik, 2310036G12Rik, 2310057K23Rik,
2310074I15Rik, AF006999, AV006427, D330041I19Rik, D830007G01Rik,
L56, mdm, shru; titin (EC:2.7.11.1); K12567 titin [EC:2.7.11.1]
Length=33467
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Query 60 GGTRCHSCV---REKVLRAFLVEEQKCVKQVLQVKE 92
GG R H V RE R++ V +KC +Q+L+V E
Sbjct 28519 GGARIHHYVIEKREASRRSWQVVSEKCTRQILKVSE 28554
Lambda K H
0.321 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2062416360
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40