bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7322_orf1
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
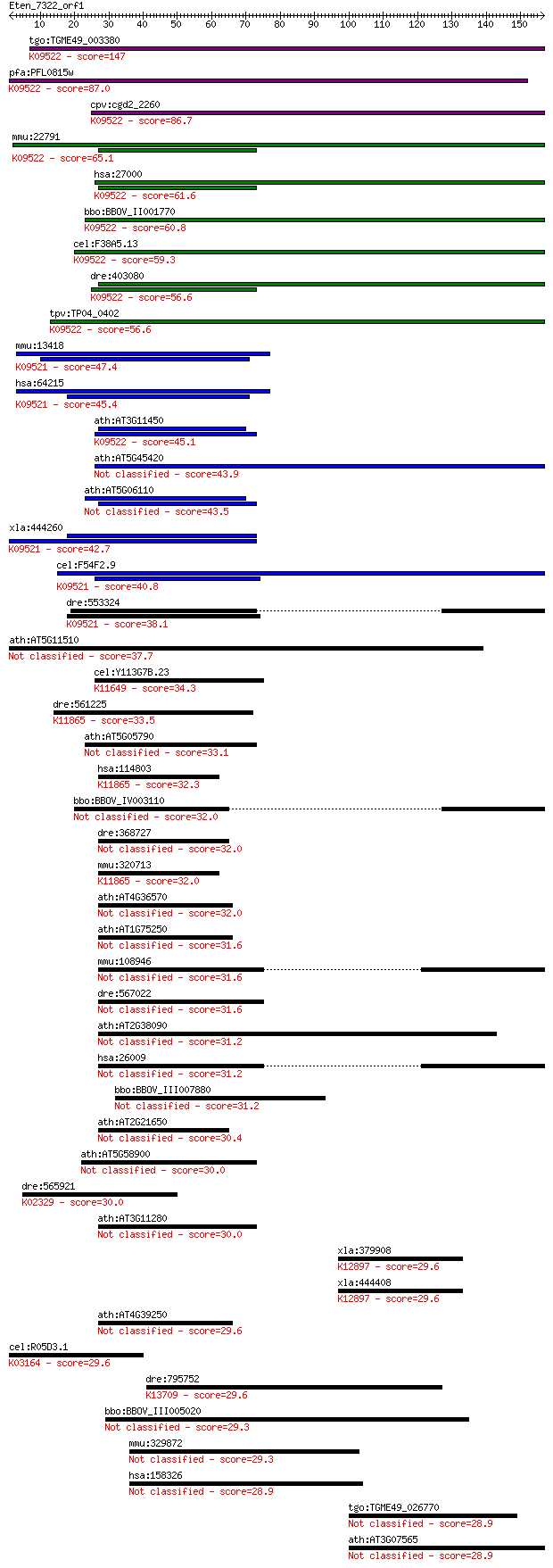
tgo:TGME49_003380 DnaJ domain-containing protein ; K09522 DnaJ... 147 1e-35
pfa:PFL0815w DNA-binding chaperone, putative; K09522 DnaJ homo... 87.0 2e-17
cpv:cgd2_2260 zuotin related factor-1 like protein with a DNAJ... 86.7 3e-17
mmu:22791 Dnajc2, AU020218, MIDA1, Zrf1, Zrf2; DnaJ (Hsp40) ho... 65.1 9e-11
hsa:27000 DNAJC2, MPHOSPH11, MPP11, ZRF1, ZUO1; DnaJ (Hsp40) h... 61.6 1e-09
bbo:BBOV_II001770 18.m06137; myb-like DNA-binding/DnaJ domain ... 60.8 2e-09
cel:F38A5.13 dnj-11; DNaJ domain (prokaryotic heat shock prote... 59.3 5e-09
dre:403080 dnajc2, zgc:85671, zrf1; DnaJ (Hsp40) homolog, subf... 56.6 3e-08
tpv:TP04_0402 DNA-binding chaperone; K09522 DnaJ homolog subfa... 56.6 3e-08
mmu:13418 Dnajc1, 4733401K02Rik, AA960110, D230036H06Rik, Dnaj... 47.4 2e-05
hsa:64215 DNAJC1, DNAJL1, ERdj1, HTJ1, MGC131954, MTJ1; DnaJ (... 45.4 7e-05
ath:AT3G11450 DNAJ heat shock N-terminal domain-containing pro... 45.1 9e-05
ath:AT5G45420 myb family transcription factor 43.9 2e-04
ath:AT5G06110 DNAJ heat shock N-terminal domain-containing pro... 43.5 3e-04
xla:444260 dnajc1, MGC80867; DnaJ (Hsp40) homolog, subfamily C... 42.7 5e-04
cel:F54F2.9 hypothetical protein; K09521 DnaJ homolog subfamil... 40.8 0.002
dre:553324 dnajc1l; zgc:152779; K09521 DnaJ homolog subfamily ... 38.1 0.012
ath:AT5G11510 MYB3R-4 (myb domain protein 3R-4); DNA binding /... 37.7 0.016
cel:Y113G7B.23 psa-1; Phasmid Socket Absent family member (psa... 34.3 0.18
dre:561225 mysm1, im:7153144, si:ch211-59d15.8; Myb-like, SWIR... 33.5 0.31
ath:AT5G05790 myb family transcription factor 33.1 0.37
hsa:114803 MYSM1, 2A-DUB, 2ADUB, DKFZp779J1554, DKFZp779J1721,... 32.3 0.62
bbo:BBOV_IV003110 21.m03046; CHY zinc finger domain containing... 32.0 0.86
dre:368727 FLJ11186, si:dz142b24.1; si:busm1-142b24.1 32.0 0.90
mmu:320713 Mysm1, C130067A03Rik, C530050H10Rik; myb-like, SWIR... 32.0 0.91
ath:AT4G36570 ATRL3; ATRL3 (ARABIDOPSIS RAD-LIKE 3); DNA bindi... 32.0 0.92
ath:AT1G75250 ATRL6; ATRL6 (ARABIDOPSIS RAD-LIKE 6); transcrip... 31.6 0.99
mmu:108946 Zzz3, 3110065C23Rik, 6430567E01Rik, AA408566, AI481... 31.6 1.2
dre:567022 wu:fj98c02; si:ch211-284g18.3 31.6 1.2
ath:AT2G38090 myb family transcription factor 31.2 1.3
hsa:26009 ZZZ3, ATAC1, DKFZp313N0119, DKFZp564I052, FLJ10362; ... 31.2 1.3
bbo:BBOV_III007880 17.m10587; hypothetical protein 31.2 1.4
ath:AT2G21650 MEE3; MEE3 (MATERNAL EFFECT EMBRYO ARREST 3); DN... 30.4 2.7
ath:AT5G58900 myb family transcription factor 30.0 2.9
dre:565921 fzd3a, fz9, fzd3, zg09; frizzled homolog 3a; K02329... 30.0 3.1
ath:AT3G11280 myb family transcription factor 30.0 3.1
xla:379908 tra2a, MGC53025; transformer 2 alpha homolog; K1289... 29.6 3.9
xla:444408 MGC82977 protein; K12897 transformer-2 protein 29.6 3.9
ath:AT4G39250 ATRL1; ATRL1 (ARABIDOPSIS RAD-LIKE 1); DNA bindi... 29.6 4.1
cel:R05D3.1 hypothetical protein; K03164 DNA topoisomerase II ... 29.6 4.2
dre:795752 arhgap5; Rho GTPase activating protein 5; K13709 Rh... 29.6 4.4
bbo:BBOV_III005020 17.m07448; hypothetical protein 29.3 4.8
mmu:329872 Frem1, BC037594, D430009N09, D630008K06, heb, qbric... 29.3 6.0
hsa:158326 FREM1, BNAR, C9orf143, C9orf145, C9orf154, FLJ25461... 28.9 6.7
tgo:TGME49_026770 cGMP-inhibited 3',5'-cyclic phosphodiesteras... 28.9 6.9
ath:AT3G07565 DNA binding 28.9 7.4
> tgo:TGME49_003380 DnaJ domain-containing protein ; K09522 DnaJ
homolog subfamily C member 2
Length=684
Score = 147 bits (372), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 85/160 (53%), Positives = 101/160 (63%), Gaps = 10/160 (6%)
Query 7 RVQEERRRQEIERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVV 66
R Q ER+ QE ++ A+Q+SWT +ELSLLAK LQKFPGGTARRW +A IGTKTQ+EVV
Sbjct 497 RRQAERKAQEEAKRLARQSSWTPDELSLLAKGLQKFPGGTARRWKLIADLIGTKTQEEVV 556
Query 67 QKAKEMSVSANLKTMGSKISQEVLEQSK-HQHA--------PASSSAARNGTQAEAEPGH 117
+K KEMS A+LK MGSKISQ +Q + H P Q A P
Sbjct 557 EKTKEMSEGASLKAMGSKISQVAFDQFRVHNQGAFKKIDADPDRKDVGETRPQTAASPAK 616
Query 118 AASVCGDSV-WTQEQQSALERALAKHPSSMPAVERWTAIA 156
+S WT QQ ALE+ALAKHP++MPA ERWTAIA
Sbjct 617 EPQETAESTDWTPAQQMALEKALAKHPATMPANERWTAIA 656
> pfa:PFL0815w DNA-binding chaperone, putative; K09522 DnaJ homolog
subfamily C member 2
Length=939
Score = 87.0 bits (214), Expect = 2e-17, Method: Composition-based stats.
Identities = 55/163 (33%), Positives = 79/163 (48%), Gaps = 16/163 (9%)
Query 1 STEEMRRVQEERRRQEIERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTK 60
ST+ + E+ QE E Q W+ +E+SLLAKAL+ +PGGT RW ++ I TK
Sbjct 747 STQSNVNIYNEQNNQENE---LQSNKWSAQEVSLLAKALKLYPGGTRNRWVLISNSIKTK 803
Query 61 TQDEVVQKAKEMSVSANLKTMGSKISQEVLEQSKHQHAPASSS------------AARNG 108
T EV++K KEM + LK +G + + K+Q+ N
Sbjct 804 TVKEVIKKTKEMFENDTLKNLGRNFDETPFDHFKNQNKGVMKKIDDNLDKREYKLTKENN 863
Query 109 TQAEAEPGHAASVCGDSVWTQEQQSALERALAKHPSSMPAVER 151
Q E + V WT E+Q LE+AL K+P+S+P ER
Sbjct 864 NQVETD-NLNGDVEKKKPWTHEEQHLLEQALIKYPTSIPIKER 905
> cpv:cgd2_2260 zuotin related factor-1 like protein with a DNAJ
domain at the N-terminus and 2 SANT domains ; K09522 DnaJ
homolog subfamily C member 2
Length=677
Score = 86.7 bits (213), Expect = 3e-17, Method: Composition-based stats.
Identities = 58/157 (36%), Positives = 86/157 (54%), Gaps = 38/157 (24%)
Query 25 ASWTVEELSLLAKALQKFPGGTARRWYHVATYI-GTKTQDEVVQKAKEMSVSANLKTMGS 83
+ WTV E+SLLAKALQK+PGG RW ++ Y+ TKT+++++ K KE+S S L +
Sbjct 507 SDWTVSEMSLLAKALQKYPGGYKNRWDMISEYLKNTKTKEQILTKVKELSESEKL----A 562
Query 84 KISQEVLEQS--------------KHQHAP----------ASSSAARNGTQAEAEPGHAA 119
K+S EV E S K + P ++SA + TQ + E
Sbjct 563 KLSNEVKEDSAFDTFIQSNKGVLKKFDNIPDVRDYSGTSIINNSAKNDATQQKKEI---- 618
Query 120 SVCGDSVWTQEQQSALERALAKHPSSMPAVERWTAIA 156
+WT++QQ +LERAL ++PSS+P+ ERW I+
Sbjct 619 -----DLWTRDQQCSLERALKQYPSSLPSNERWELIS 650
> mmu:22791 Dnajc2, AU020218, MIDA1, Zrf1, Zrf2; DnaJ (Hsp40)
homolog, subfamily C, member 2; K09522 DnaJ homolog subfamily
C member 2
Length=621
Score = 65.1 bits (157), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 52/171 (30%), Positives = 84/171 (49%), Gaps = 24/171 (14%)
Query 2 TEEMRRVQEE---RRRQ-------EIERKKAQQASWTVEELSLLAKALQKFPGGTARRWY 51
E+MRR +EE R RQ + +W+ ++L LL KA+ FP GT RW
Sbjct 419 NEQMRREKEEADARMRQASKNAEKSTGGSGSGSKNWSEDDLQLLIKAVNLFPAGTNSRWE 478
Query 52 HVATYIGTKTQDEVVQKAKE-MSVSANLKTMG----SKISQEVLEQSKHQHAPASSSAAR 106
+A Y+ + V + AK+ +S + +L+ + I+++ ++ K +H AS + +
Sbjct 479 VIANYMNIHSSSGVKRTAKDVISKAKSLQKLDPHQKDDINKKAFDKFKKEHGVASQADSA 538
Query 107 NGTQAEAEPGHAASVCGDSV-WTQEQQSALERALAKHPSSMPAVERWTAIA 156
++ P C DS WT E+Q LE+AL +P + P ERW IA
Sbjct 539 APSERFEGP------CIDSTPWTTEEQKLLEQALKTYPVNTP--ERWEKIA 581
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
WT EE LL +AL+ +P T RW +A + +T+ + +++ KE+
Sbjct 554 WTTEEQKLLEQALKTYPVNTPERWEKIAEAVPGRTKKDCMRRYKEL 599
> hsa:27000 DNAJC2, MPHOSPH11, MPP11, ZRF1, ZUO1; DnaJ (Hsp40)
homolog, subfamily C, member 2; K09522 DnaJ homolog subfamily
C member 2
Length=568
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 42/136 (30%), Positives = 68/136 (50%), Gaps = 12/136 (8%)
Query 26 SWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMSVSA-NLKTMG-- 82
+W+ ++L LL KA+ FP GT RW +A Y+ + V + AK++ A +L+ +
Sbjct 400 NWSEDDLQLLIKAVNLFPAGTNSRWEVIANYMNIHSSSGVKRTAKDVIGKAKSLQKLDPH 459
Query 83 --SKISQEVLEQSKHQHAPASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQSALERALA 140
I+++ ++ K +H A N T +E G + WT E+Q LE+AL
Sbjct 460 QKDDINKKAFDKFKKEHGVVPQ--ADNATPSERFEGPYTDF---TPWTTEEQKLLEQALK 514
Query 141 KHPSSMPAVERWTAIA 156
+P + P ERW IA
Sbjct 515 TYPVNTP--ERWEKIA 528
Score = 38.5 bits (88), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 28/46 (60%), Gaps = 0/46 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
WT EE LL +AL+ +P T RW +A + +T+ + +++ KE+
Sbjct 501 WTTEEQKLLEQALKTYPVNTPERWEKIAEAVPGRTKKDCMKRYKEL 546
> bbo:BBOV_II001770 18.m06137; myb-like DNA-binding/DnaJ domain
containing protein; K09522 DnaJ homolog subfamily C member
2
Length=647
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 41/134 (30%), Positives = 61/134 (45%), Gaps = 31/134 (23%)
Query 23 QQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMSVSANLKTMG 82
Q WT EELS L+K ++ G RW +A ++ TKT + +Q A+E++ L
Sbjct 516 QPVQWTTEELSRLSKGVEMHVAGVTDRWSLIAKHVKTKTAAQCIQMAREIASGKRLD--- 572
Query 83 SKISQEVLEQSKHQHAPASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQSALERALAKH 142
++ PA + A NG +++ W+ EQQS E AL K+
Sbjct 573 -------------ENTPAVNIA--NGVHSDS-------------WSVEQQSEFEAALVKY 604
Query 143 PSSMPAVERWTAIA 156
PSS+ RW IA
Sbjct 605 PSSLDPASRWRLIA 618
> cel:F38A5.13 dnj-11; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-11); K09522 DnaJ homolog subfamily
C member 2
Length=589
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 45/137 (32%), Positives = 63/137 (45%), Gaps = 22/137 (16%)
Query 20 KKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMSVSANLK 79
K+A + +WT EE+ LL KA FP GT RW +A YI +D S K
Sbjct 449 KQADKETWTSEEIQLLVKASNTFPPGTVERWVQIADYINEHRKD---------STGLPPK 499
Query 80 TMGSKISQEVLEQSKHQHAPASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQSALERAL 139
T I Q Q+ + P S++ + GT ++ + VW+ +Q LE A+
Sbjct 500 TEKQVIKQCKAVQTMNVKLP-STTQNQLGT----------ALPDEDVWSATEQKTLEDAI 548
Query 140 AKHPSSMPAVERWTAIA 156
KH SS P ERW I+
Sbjct 549 KKHKSSDP--ERWEKIS 563
> dre:403080 dnajc2, zgc:85671, zrf1; DnaJ (Hsp40) homolog, subfamily
C, member 2; K09522 DnaJ homolog subfamily C member
2
Length=618
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 45/141 (31%), Positives = 69/141 (48%), Gaps = 21/141 (14%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGT-------KTQDEVVQKAKEM-SVSANL 78
W+ E+L LL KA+ FP GT RW +A Y+ +T +V+ KAK + + +
Sbjct 450 WSEEDLQLLIKAVNLFPAGTNARWEVIANYMNQHSSSGVRRTAKDVINKAKTLQKLDPHQ 509
Query 79 KTMGSKISQEVLEQSKHQHAPASSSAARNGTQAEAEPGHAASVCGDS---VWTQEQQSAL 135
K +I+++ E+ K +H+ A N +E +V DS WT E+Q L
Sbjct 510 K---DEINRKAFEKFKKEHS-AVPPTVDNAMPSE----RFDAVGADSNAAAWTTEEQKLL 561
Query 136 ERALAKHPSSMPAVERWTAIA 156
E+AL +P S ERW I+
Sbjct 562 EQALKTYPVS--TAERWERIS 580
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 31/48 (64%), Gaps = 0/48 (0%)
Query 25 ASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
A+WT EE LL +AL+ +P TA RW ++ + +++ + +++ KE+
Sbjct 551 AAWTTEEQKLLEQALKTYPVSTAERWERISEAVPGRSKKDCMKRYKEL 598
> tpv:TP04_0402 DNA-binding chaperone; K09522 DnaJ homolog subfamily
C member 2
Length=674
Score = 56.6 bits (135), Expect = 3e-08, Method: Composition-based stats.
Identities = 40/144 (27%), Positives = 65/144 (45%), Gaps = 27/144 (18%)
Query 13 RRQEIERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
R + + + WT E+L L+K ++ P GT RW ++ Y+ TKT + ++ +K +
Sbjct 529 RAESVGTTPSAVTEWTKEDLKRLSKGVEINPAGTPGRWNLISKYVKTKTAAQCIEMSKLI 588
Query 73 SVSANLKTMGSKISQEVLEQSKHQHAPASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQ 132
S + T G+ +S + NGT + S W++ QQ
Sbjct 589 SNNITDNTTGT-----------------TSVHSTNGTDTVSM----------SSWSELQQ 621
Query 133 SALERALAKHPSSMPAVERWTAIA 156
+A E AL K+PS + V RW IA
Sbjct 622 TAFESALKKYPSHLDPVVRWRKIA 645
> mmu:13418 Dnajc1, 4733401K02Rik, AA960110, D230036H06Rik, Dnajl1,
ERdj1, ERj1p, MTJ1; DnaJ (Hsp40) homolog, subfamily C,
member 1; K09521 DnaJ homolog subfamily C member 1
Length=552
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/74 (40%), Positives = 40/74 (54%), Gaps = 5/74 (6%)
Query 3 EEMRRVQEERRRQEIERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQ 62
E+M E R+R + K Q WT E+LS L +++ KFPGGT RW +A +G
Sbjct 308 EQMDDWLENRKRTQ----KRQAPEWTEEDLSQLTRSMVKFPGGTPGRWDKIAHELGRSVT 363
Query 63 DEVVQKAKEMSVSA 76
D V KAKE+ S
Sbjct 364 D-VTTKAKELKDSV 376
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 39/61 (63%), Gaps = 2/61 (3%)
Query 10 EERRRQEIERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKA 69
EE++R+E R +A + +WT + LL ALQ++P G + RW +A + +K++++ + +
Sbjct 480 EEKQRKE--RTRAAEEAWTQSQQKLLELALQQYPKGASDRWDKIAKCVPSKSKEDCIARY 537
Query 70 K 70
K
Sbjct 538 K 538
> hsa:64215 DNAJC1, DNAJL1, ERdj1, HTJ1, MGC131954, MTJ1; DnaJ
(Hsp40) homolog, subfamily C, member 1; K09521 DnaJ homolog
subfamily C member 1
Length=554
Score = 45.4 bits (106), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 29/74 (39%), Positives = 39/74 (52%), Gaps = 5/74 (6%)
Query 3 EEMRRVQEERRRQEIERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQ 62
E+M E R R + K Q WT E+LS L +++ KFPGGT RW +A +G
Sbjct 310 EQMDDWLENRNRTQ----KKQAPEWTEEDLSQLTRSMVKFPGGTPGRWEKIAHELGRSVT 365
Query 63 DEVVQKAKEMSVSA 76
D V KAK++ S
Sbjct 366 D-VTTKAKQLKDSV 378
Score = 37.7 bits (86), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 33/53 (62%), Gaps = 0/53 (0%)
Query 18 ERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAK 70
ER ++ + WT + LL ALQ++P G++ RW +A + +K++++ + + K
Sbjct 488 ERARSAEEPWTQNQQKLLELALQQYPRGSSDRWDKIARCVPSKSKEDCIARYK 540
> ath:AT3G11450 DNAJ heat shock N-terminal domain-containing protein
/ cell division protein-related; K09522 DnaJ homolog
subfamily C member 2
Length=663
Score = 45.1 bits (105), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKA 69
W+ EE+ +L K + K+P GT+RRW ++ YIGT E + KA
Sbjct 482 WSKEEIDMLRKGMIKYPKGTSRRWEVISEYIGTGRSVEEILKA 524
Score = 32.3 bits (72), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 26 SWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
SW+ + L +AL+ FP T++RW VA + KT ++ +K E+
Sbjct 606 SWSTVQERALVQALKTFPKETSQRWERVAAAVPGKTMNQCKKKFAEL 652
> ath:AT5G45420 myb family transcription factor
Length=309
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 41/133 (30%), Positives = 56/133 (42%), Gaps = 14/133 (10%)
Query 26 SWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDE-VVQKAKEMSVSANLKTMGSK 84
WT EE+ +L K L K P G RW VA+ G + + E V++KAKE+ K S
Sbjct 159 DWTAEEIEILKKQLIKHPAGKPGRWETVASAFGGRYKTENVIKKAKEI---GEKKIYESD 215
Query 85 ISQEVLEQSKHQHAPASSSAARNGTQAEAEPGHAASVCG-DSVWTQEQQSALERALAKHP 143
+ L+ K +S R + E G G +W+ + AL AL P
Sbjct 216 DYAQFLKNRK-------ASDPRLVDENEENSGAGGDAEGTKEIWSNGEDIALLNALKAFP 268
Query 144 SSMPAVERWTAIA 156
A RW IA
Sbjct 269 KE--AAMRWEKIA 279
> ath:AT5G06110 DNAJ heat shock N-terminal domain-containing protein
/ cell division protein-related
Length=663
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 23 QQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKA 69
++ W+ EE+ +L K KFP GT++RW ++ YIGT E + KA
Sbjct 473 KEKPWSKEEIDMLRKGTTKFPKGTSQRWEVISEYIGTGRSVEEILKA 519
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
W+ + L +AL+ FP T +RW VAT + KT ++ +K ++
Sbjct 607 WSAVQERALVQALKTFPKETNQRWERVATAVPGKTMNQCKKKFADL 652
> xla:444260 dnajc1, MGC80867; DnaJ (Hsp40) homolog, subfamily
C, member 1; K09521 DnaJ homolog subfamily C member 1
Length=534
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/55 (43%), Positives = 34/55 (61%), Gaps = 3/55 (5%)
Query 18 ERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
++KKA + WT E+LS L + + KFPGGT RW +A +G D V KAK++
Sbjct 305 QKKKAPE--WTEEDLSQLTRNMTKFPGGTPGRWEKIAHELGRSVTD-VTTKAKQL 356
Score = 35.4 bits (80), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 38/72 (52%), Gaps = 9/72 (12%)
Query 1 STEEMRRVQEERRRQEIERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTK 60
S +MR+ ++ R +++ W+ + LL ALQ++P GT RW +A + K
Sbjct 460 SDNDMRKKEQSRSSEDL---------WSQNQQKLLELALQQYPKGTGERWDKIAKCVPGK 510
Query 61 TQDEVVQKAKEM 72
++++ + + K +
Sbjct 511 SKEDCICRYKLL 522
> cel:F54F2.9 hypothetical protein; K09521 DnaJ homolog subfamily
C member 1
Length=414
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/143 (23%), Positives = 60/143 (41%), Gaps = 29/143 (20%)
Query 15 QEIERKKAQQASWTVEEL-SLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMS 73
E+ +K A+WT +EL SL+ + +K+P GT RW + + +D + +
Sbjct 270 NEVVAQKQSGATWTPDELASLVRLSTEKYPAGTPNRWEQMGRVLNRSAEDVI-------A 322
Query 74 VSANLKTMGSKISQEVLEQSKHQHAPASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQS 133
++ +K M + ++L + Q P + + W+Q +Q
Sbjct 323 MAGKMKQMKQEDYTKLLMTTIQQSVPVEEKSEDD-------------------WSQAEQK 363
Query 134 ALERALAKHPSSMPAVERWTAIA 156
A E AL K+P ERW I+
Sbjct 364 AFETALQKYPKGTD--ERWERIS 384
Score = 37.0 bits (84), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 29/48 (60%), Gaps = 0/48 (0%)
Query 26 SWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMS 73
W+ E ALQK+P GT RW ++ IG+KT+ +V+ + K+++
Sbjct 356 DWSQAEQKAFETALQKYPKGTDERWERISEEIGSKTKKQVMVRFKQLA 403
> dre:553324 dnajc1l; zgc:152779; K09521 DnaJ homolog subfamily
C member 1
Length=526
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 34/54 (62%), Gaps = 3/54 (5%)
Query 19 RKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
+KK Q+ WT ++ S L++ + KFPGGT RW +A +G ++ EV K K++
Sbjct 302 KKKTQE--WTEDDHSQLSRCMAKFPGGTPGRWEKIAHELG-RSVSEVTAKVKQI 352
Score = 37.4 bits (85), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 18 ERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMS 73
E+ A + WT + LL ALQ++P GT RW +A + KT++E + + K ++
Sbjct 460 EKAGAAEDVWTQNQQRLLELALQQYPRGTTERWDKIAKVVPGKTKEECMCRFKLLA 515
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 16/30 (53%), Gaps = 2/30 (6%)
Query 127 WTQEQQSALERALAKHPSSMPAVERWTAIA 156
WT++ S L R +AK P P RW IA
Sbjct 308 WTEDDHSQLSRCMAKFPGGTPG--RWEKIA 335
> ath:AT5G11510 MYB3R-4 (myb domain protein 3R-4); DNA binding
/ transcription coactivator/ transcription factor
Length=798
Score = 37.7 bits (86), Expect = 0.016, Method: Composition-based stats.
Identities = 35/145 (24%), Positives = 60/145 (41%), Gaps = 10/145 (6%)
Query 1 STEEMRRVQEERRRQEIERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTK 60
ST + R + R +++ + WT EE +L KA+ F G + W +A Y +
Sbjct 6 STPQERIPKLRHGRTSGPARRSTRGQWTAEEDEILRKAVHSFKG---KNWKKIAEYFKDR 62
Query 61 TQDEVVQKAKEMSVSANLKTMGSKISQEVLEQSKHQHAPASSSA------ARNGTQA-EA 113
T + + + +++ +K +K E++ Q ++ P S R G Q E
Sbjct 63 TDVQCLHRWQKVLNPELVKGPWTKEEDEMIVQLIEKYGPKKWSTIARFLPGRIGKQCRER 122
Query 114 EPGHAASVCGDSVWTQEQQSALERA 138
H WTQE++ L RA
Sbjct 123 WHNHLNPAINKEAWTQEEELLLIRA 147
> cel:Y113G7B.23 psa-1; Phasmid Socket Absent family member (psa-1);
K11649 SWI/SNF related-matrix-associated actin-dependent
regulator of chromatin subfamily C
Length=789
Score = 34.3 bits (77), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 4/49 (8%)
Query 26 SWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMSV 74
WT +E LL +AL+ F W V ++GT+TQ E V K ++ +
Sbjct 256 DWTEQETCLLLEALEMF----KDDWNKVCDHVGTRTQHECVLKFLQLPI 300
> dre:561225 mysm1, im:7153144, si:ch211-59d15.8; Myb-like, SWIRM
and MPN domains 1 (EC:3.1.2.15); K11865 protein MYSM1
Length=822
Score = 33.5 bits (75), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 29/58 (50%), Gaps = 4/58 (6%)
Query 14 RQEIERKKAQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKE 71
++ + + A Q W EE L K L +F RRW +A IGT+T +V AK+
Sbjct 89 KKPLVKSSAAQTRWAEEEKELFEKGLAQF----GRRWTKIAKLIGTRTVLQVKSYAKQ 142
> ath:AT5G05790 myb family transcription factor
Length=277
Score = 33.1 bits (74), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 23 QQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
Q +SWT EE +AL + T RW+ VA I KT +V+++ ++
Sbjct 27 QSSSWTKEENKKFERALAVYADDTPDRWFKVAAMIPGKTISDVMRQYSKL 76
> hsa:114803 MYSM1, 2A-DUB, 2ADUB, DKFZp779J1554, DKFZp779J1721,
KIAA1915, RP4-592A1.1; Myb-like, SWIRM and MPN domains 1
(EC:3.1.2.15); K11865 protein MYSM1
Length=828
Score = 32.3 bits (72), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 4/35 (11%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKT 61
WT+EE L + L KF RRW ++ IG++T
Sbjct 121 WTIEEKELFEQGLAKF----GRRWTKISKLIGSRT 151
> bbo:BBOV_IV003110 21.m03046; CHY zinc finger domain containing
protein
Length=549
Score = 32.0 bits (71), Expect = 0.86, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 127 WTQEQQSALERALAKHPSSMPAVERWTAIA 156
W+ E+Q +LE AL K A ERW+A+A
Sbjct 177 WSLEEQKSLEFALGKTKGIPNAAERWSAVA 206
Score = 30.0 bits (66), Expect = 3.1, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 2/47 (4%)
Query 20 KKAQQASWTVEELSLLAKALQKFPG--GTARRWYHVATYIGTKTQDE 64
+++ +W++EE L AL K G A RW VA + TK+ DE
Sbjct 170 RESNVDTWSLEEQKSLEFALGKTKGIPNAAERWSAVAAIVKTKSADE 216
> dre:368727 FLJ11186, si:dz142b24.1; si:busm1-142b24.1
Length=1010
Score = 32.0 bits (71), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDE 64
WT EEL L + + P + W HVA +GT++ +E
Sbjct 856 WTEEELQKLHEVVNSLPKHQSGYWAHVAYVVGTRSAEE 893
> mmu:320713 Mysm1, C130067A03Rik, C530050H10Rik; myb-like, SWIRM
and MPN domains 1 (EC:3.1.2.15); K11865 protein MYSM1
Length=819
Score = 32.0 bits (71), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 20/35 (57%), Gaps = 4/35 (11%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKT 61
WTVEE L + L KF RRW +AT + ++T
Sbjct 118 WTVEEKELFEQGLAKF----GRRWTKIATLLKSRT 148
> ath:AT4G36570 ATRL3; ATRL3 (ARABIDOPSIS RAD-LIKE 3); DNA binding
/ transcription factor
Length=58
Score = 32.0 bits (71), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEV 65
WT +E L +AL + T RW++VA +G K+ +EV
Sbjct 12 WTRKENKLFERALATYDQDTPDRWHNVARAVGGKSAEEV 50
> ath:AT1G75250 ATRL6; ATRL6 (ARABIDOPSIS RAD-LIKE 6); transcription
factor
Length=97
Score = 31.6 bits (70), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEV 65
WT + + +AL + T RW++VA +G KT +EV
Sbjct 12 WTFSQNKMFERALAVYDKDTPDRWHNVAKAVGGKTVEEV 50
> mmu:108946 Zzz3, 3110065C23Rik, 6430567E01Rik, AA408566, AI481057,
AW556059, R75514; zinc finger, ZZ domain containing 3
Length=910
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Query 27 WTVEELSLLAKALQKFPGG--TARRWYHVATYIGTKTQDEVVQKAKEMSV 74
WTVEE L + L K+P +RRW +A +G +T +V + ++ +
Sbjct 662 WTVEEQKKLEQLLLKYPPEEVESRRWQKIADELGNRTAKQVASRVQKYFI 711
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 7/43 (16%)
Query 121 VCGDS-------VWTQEQQSALERALAKHPSSMPAVERWTAIA 156
+C DS +WT E+Q LE+ L K+P RW IA
Sbjct 649 LCDDSKPETFNQLWTVEEQKKLEQLLLKYPPEEVESRRWQKIA 691
> dre:567022 wu:fj98c02; si:ch211-284g18.3
Length=871
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Query 27 WTVEELSLLAKALQKFPGG--TARRWYHVATYIGTKTQDEVVQKAKEMSV 74
WT+EE L + L KFP ++RW +A +G +T +V + ++ +
Sbjct 624 WTIEEQKKLEQLLLKFPPEEVESKRWQKIADELGNRTAKQVASRVQKYFI 673
> ath:AT2G38090 myb family transcription factor
Length=298
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 30/130 (23%), Positives = 48/130 (36%), Gaps = 14/130 (10%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM-----SVSANLKTM 81
WT EE AL + T RW VA + KT +V+++ +E+ + A L +
Sbjct 29 WTAEENKKFENALAFYDKDTPDRWSRVAAMLPGKTVGDVIKQYRELEEDVSDIEAGLIPI 88
Query 82 GSKISQEV-LEQSKHQHAPASSSAARNGTQAEAEPGHAASVCGDS--------VWTQEQQ 132
S L+ + A ++ NG A G S + WT+E+
Sbjct 89 PGYASDSFTLDWGGYDGASGNNGFNMNGYYFSAAGGKRGSAARTAEHERKKGVPWTEEEH 148
Query 133 SALERALAKH 142
L K+
Sbjct 149 RQFLMGLKKY 158
> hsa:26009 ZZZ3, ATAC1, DKFZp313N0119, DKFZp564I052, FLJ10362;
zinc finger, ZZ-type containing 3
Length=903
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Query 27 WTVEELSLLAKALQKFPGG--TARRWYHVATYIGTKTQDEVVQKAKEMSV 74
WTVEE L + L K+P +RRW +A +G +T +V + ++ +
Sbjct 655 WTVEEQKKLEQLLIKYPPEEVESRRWQKIADELGNRTAKQVASRVQKYFI 704
Score = 28.5 bits (62), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 21/43 (48%), Gaps = 7/43 (16%)
Query 121 VCGDS-------VWTQEQQSALERALAKHPSSMPAVERWTAIA 156
+C D+ +WT E+Q LE+ L K+P RW IA
Sbjct 642 LCDDTKPETFNQLWTVEEQKKLEQLLIKYPPEEVESRRWQKIA 684
> bbo:BBOV_III007880 17.m10587; hypothetical protein
Length=1139
Score = 31.2 bits (69), Expect = 1.4, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 28/61 (45%), Gaps = 0/61 (0%)
Query 32 LSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMSVSANLKTMGSKISQEVLE 91
+ LL+ F RW ++ TQ+ V AK S +AN K SK +EVL+
Sbjct 122 VKLLSNLWDGFESSGVDRWITNNMWLQLLTQEYQVHTAKHNSANANRKYSNSKRVEEVLQ 181
Query 92 Q 92
Q
Sbjct 182 Q 182
> ath:AT2G21650 MEE3; MEE3 (MATERNAL EFFECT EMBRYO ARREST 3);
DNA binding / transcription factor
Length=101
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDE 64
WTV++ +AL + T RW++VA +G KT +E
Sbjct 14 WTVKQNKAFERALAVYDQDTPDRWHNVARAVGGKTPEE 51
> ath:AT5G58900 myb family transcription factor
Length=288
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 22 AQQASWTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
+ A+WT E AL + T RW VA I KT +V+++ ++
Sbjct 29 GEGATWTAAENKAFENALAVYDDNTPDRWQKVAAVIPGKTVSDVIRQYNDL 79
> dre:565921 fzd3a, fz9, fzd3, zg09; frizzled homolog 3a; K02329
frizzled 3
Length=685
Score = 30.0 bits (66), Expect = 3.1, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 5 MRRVQEERRRQEIERKKAQQASWTVEELSLLAKALQKFPGGTARR 49
+ R+ + R ++R ++Q +V +LS+ A+A+Q PG R
Sbjct 632 INRLDSQSRHGSLQRLESQSRHSSVRDLSITAQAIQSSPGNGIHR 676
> ath:AT3G11280 myb family transcription factor
Length=263
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEM 72
WT EE + +AL + + RW+ VA+ I KT +V+++ ++
Sbjct 33 WTKEENKMFERALAIYAEDSPDRWFKVASMIPGKTVFDVMKQYSKL 78
> xla:379908 tra2a, MGC53025; transformer 2 alpha homolog; K12897
transformer-2 protein
Length=276
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 97 HAPASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQ 132
H+P S+ NG++A +P V G S++T E+
Sbjct 92 HSPMSNRRRHNGSRANPDPNLCIGVFGLSLYTTERD 127
> xla:444408 MGC82977 protein; K12897 transformer-2 protein
Length=276
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 97 HAPASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQ 132
H+P S+ NG++A +P V G S++T E+
Sbjct 92 HSPMSNRRRHNGSRANPDPNICVGVFGLSLYTTERD 127
> ath:AT4G39250 ATRL1; ATRL1 (ARABIDOPSIS RAD-LIKE 1); DNA binding
/ transcription factor
Length=100
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 27 WTVEELSLLAKALQKFPGGTARRWYHVATYIGTKTQDEV 65
WT ++ +AL + T RW +VA +G KT +EV
Sbjct 14 WTAKQNKAFEQALATYDQDTPNRWQNVAKVVGGKTTEEV 52
> cel:R05D3.1 hypothetical protein; K03164 DNA topoisomerase II
[EC:5.99.1.3]
Length=1054
Score = 29.6 bits (65), Expect = 4.2, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 26/42 (61%), Gaps = 3/42 (7%)
Query 1 STEEMRRVQEERRRQEIERKKAQQASWTV---EELSLLAKAL 39
+++EM+R++E + R+ E + A+ A W EL LA+A+
Sbjct 1007 TSDEMKRLEERKSRRRTELEAAESADWKSVWRSELDKLAEAV 1048
> dre:795752 arhgap5; Rho GTPase activating protein 5; K13709
Rho GTPase-activating protein 5
Length=1502
Score = 29.6 bits (65), Expect = 4.4, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 37/88 (42%), Gaps = 4/88 (4%)
Query 41 KFPGGTARRWYHVATYIGTKTQDEVVQKAKEMSVSANLKTM--GSKISQEVLEQSKHQHA 98
KF + YH T + T +EV++K + S G+ +++V QS H H+
Sbjct 913 KFTALYSLSQYHRQTEVFTPFFNEVLEKKSNIESSFMFDNSRDGTGTTEDVFPQSPHSHS 972
Query 99 PASSSAARNGTQAEAEPGHAASVCGDSV 126
PA + E P + S GD V
Sbjct 973 PAYQYYPDSEDDGEGPPPY--SPIGDDV 998
> bbo:BBOV_III005020 17.m07448; hypothetical protein
Length=752
Score = 29.3 bits (64), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 38/119 (31%), Positives = 53/119 (44%), Gaps = 28/119 (23%)
Query 29 VEELSL-LAKALQKFPGGTARRWYHVATY---IGTKTQD---------EVVQKAKEMSVS 75
+E++ L L KA K+ + H TY + KT D EVV+KAKEM
Sbjct 185 LEDMRLELKKANDKYEEASETLKAHEITYKKVLDAKTSDLDAAKSQQKEVVEKAKEMLAE 244
Query 76 ANLKTMGSKISQEVLEQSKHQHAPASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQSA 134
A QE LEQ + +H A+ N AE + +AA D+V T+ Q+ A
Sbjct 245 A----------QETLEQKQKEHEEATI----NFNNAE-KVVNAAMEGNDAVNTKTQELA 288
> mmu:329872 Frem1, BC037594, D430009N09, D630008K06, heb, qbrick;
Fras1 related extracellular matrix protein 1
Length=2172
Score = 29.3 bits (64), Expect = 6.0, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Query 36 AKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMSVSANLKTMGSKISQEVLEQSKH 95
+K LQ PG + + W TY G + DEV + V+A L T +K + ++L+
Sbjct 1794 SKLLQFDPGMSTKMWNIAITYDGLEEDDEVFEVILNSPVNAVLGTQ-TKAAVKILDSKGG 1852
Query 96 QHAPASS 102
+ P++S
Sbjct 1853 RCHPSNS 1859
> hsa:158326 FREM1, BNAR, C9orf143, C9orf145, C9orf154, FLJ25461,
TILRR; FRAS1 related extracellular matrix 1
Length=715
Score = 28.9 bits (63), Expect = 6.7, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 34/68 (50%), Gaps = 1/68 (1%)
Query 36 AKALQKFPGGTARRWYHVATYIGTKTQDEVVQKAKEMSVSANLKTMGSKISQEVLEQSKH 95
+K +Q PG + + W TY G + DEV + V+A L T +K + ++L+
Sbjct 331 SKLIQFDPGMSTKMWNIAITYDGLEEDDEVFEVILNSPVNAVLGTK-TKAAVKILDSKGG 389
Query 96 QHAPASSS 103
Q P+ SS
Sbjct 390 QCHPSYSS 397
> tgo:TGME49_026770 cGMP-inhibited 3',5'-cyclic phosphodiesterase,
putative (EC:3.1.4.17)
Length=995
Score = 28.9 bits (63), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 20/49 (40%), Positives = 23/49 (46%), Gaps = 7/49 (14%)
Query 100 ASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQSALERALAKHPSSMPA 148
ASS +G A PG SVCG S WTQ R L +H + PA
Sbjct 709 ASSPPHAHGPGASTPPG-PQSVCGASFWTQ------RRLLVRHKTYSPA 750
> ath:AT3G07565 DNA binding
Length=192
Score = 28.9 bits (63), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 3/57 (5%)
Query 100 ASSSAARNGTQAEAEPGHAASVCGDSVWTQEQQSALERALAKHPSSMPAVERWTAIA 156
++ AA + +Q H + D WT E+QS LE L K+ ++ P+V R+ IA
Sbjct 41 GNTGAAADNSQTIGALRHNPGISTD--WTLEEQSLLEDLLVKY-ATEPSVFRYAKIA 94
Lambda K H
0.308 0.117 0.326
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3451799900
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40