bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7301_orf1
Length=161
Score E
Sequences producing significant alignments: (Bits) Value
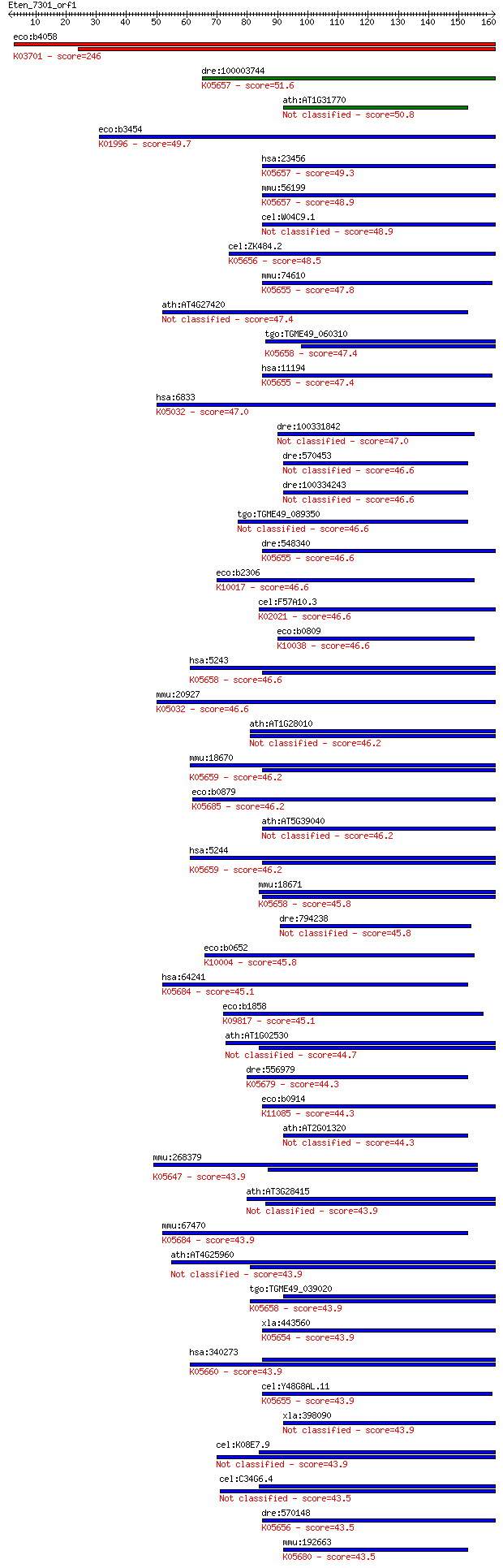
eco:b4058 uvrA, dar, ECK4050, JW4019; ATPase and DNA damage re... 246 3e-65
dre:100003744 abcb10, si:dkey-162b3.3; ATP-binding cassette, s... 51.6 1e-06
ath:AT1G31770 ABC transporter family protein 50.8 2e-06
eco:b3454 livF, ECK3438, JW3419; leucine/isoleucine/valine tra... 49.7 4e-06
hsa:23456 ABCB10, EST20237, M-ABC2, MTABC2; ATP-binding casset... 49.3 6e-06
mmu:56199 Abcb10, AU016331, Abc-me, Abcb12, C76534; ATP-bindin... 48.9 7e-06
cel:W04C9.1 haf-4; HAlF transporter (PGP related) family membe... 48.9 8e-06
cel:ZK484.2 haf-9; HAlF transporter (PGP related) family membe... 48.5 9e-06
mmu:74610 Abcb8, 4833412N02Rik, AA409895; ATP-binding cassette... 47.8 1e-05
ath:AT4G27420 ABC transporter family protein 47.4 2e-05
tgo:TGME49_060310 ATP-binding cassette protein subfamily B mem... 47.4 2e-05
hsa:11194 ABCB8, EST328128, M-ABC1, MABC1; ATP-binding cassett... 47.4 2e-05
hsa:6833 ABCC8, ABC36, HHF1, HI, HRINS, MRP8, PHHI, SUR, SUR1,... 47.0 2e-05
dre:100331842 hypothetical protein LOC100331842 47.0 3e-05
dre:570453 abcg2b; ATP-binding cassette, sub-family G (WHITE),... 46.6 3e-05
dre:100334243 ATP-binding cassette, sub-family G (WHITE), memb... 46.6 3e-05
tgo:TGME49_089350 white protein, putative (EC:3.6.3.28) 46.6 3e-05
dre:548340 abcb8, zgc:113037; ATP-binding cassette, sub-family... 46.6 3e-05
eco:b2306 hisP, ECK2300, JW2303; histidine/lysine/arginine/orn... 46.6 4e-05
cel:F57A10.3 haf-3; HAlF transporter (PGP related) family memb... 46.6 4e-05
eco:b0809 glnQ, ECK0798, JW0794; glutamine transporter subunit... 46.6 4e-05
hsa:5243 ABCB1, ABC20, CD243, CLCS, GP170, MDR1, MGC163296, P-... 46.6 4e-05
mmu:20927 Abcc8, D930031B21Rik, SUR1, Sur; ATP-binding cassett... 46.6 4e-05
ath:AT1G28010 PGP14; PGP14 (P-GLYCOPROTEIN 14); ATPase, couple... 46.2 4e-05
mmu:18670 Abcb4, Mdr2, Pgy-2, Pgy2, mdr-2; ATP-binding cassett... 46.2 4e-05
eco:b0879 macB, ECK0870, JW0863, ybjZ; fused macrolide transpo... 46.2 5e-05
ath:AT5G39040 ATTAP2; ATTAP2; ATPase, coupled to transmembrane... 46.2 5e-05
hsa:5244 ABCB4, ABC21, GBD1, MDR2, MDR2/3, MDR3, PFIC-3, PGY3;... 46.2 5e-05
mmu:18671 Abcb1a, Abcb4, Evi32, Mdr1a, Mdr3, P-gp, Pgp, Pgy-3,... 45.8 6e-05
dre:794238 abcg4b, zgc:171697; ATP-binding cassette, sub-famil... 45.8 6e-05
eco:b0652 gltL, ECK0645, JW0647; glutamate and aspartate trans... 45.8 7e-05
hsa:64241 ABCG8, GBD4, MGC142217, STSL; ATP-binding cassette, ... 45.1 1e-04
eco:b1858 znuC, ECK1859, JW1847, yebM; zinc transporter subuni... 45.1 1e-04
ath:AT1G02530 PGP12; PGP12 (P-GLYCOPROTEIN 12); ATPase, couple... 44.7 1e-04
dre:556979 abcg1, zgc:162197; ATP-binding cassette, sub-family... 44.3 2e-04
eco:b0914 msbA, ECK0905, JW0897; fused lipid transporter subun... 44.3 2e-04
ath:AT2G01320 ABC transporter family protein 44.3 2e-04
mmu:268379 Abca13, 9830132L24, A930002G16Rik, AI956815; ATP-bi... 43.9 2e-04
ath:AT3G28415 P-glycoprotein, putative 43.9 2e-04
mmu:67470 Abcg8, 1300003C16Rik, AI114946, sterolin-2; ATP-bind... 43.9 2e-04
ath:AT4G25960 PGP2; PGP2 (P-GLYCOPROTEIN 2); ATPase, coupled t... 43.9 2e-04
tgo:TGME49_039020 ATP-binding cassette protein subfamily B mem... 43.9 2e-04
xla:443560 tap2, abc18, abcb3, apt2, psf2, ring11; transporter... 43.9 2e-04
hsa:340273 ABCB5, ABCB5alpha, ABCB5beta, EST422562; ATP-bindin... 43.9 2e-04
cel:Y48G8AL.11 haf-6; HAlF transporter (PGP related) family me... 43.9 2e-04
xla:398090 TAP2 protein 43.9 2e-04
cel:K08E7.9 pgp-1; P-GlycoProtein related family member (pgp-1) 43.9 3e-04
cel:C34G6.4 pgp-2; P-GlycoProtein related family member (pgp-2) 43.5 3e-04
dre:570148 abcb9, si:dkey-21k4.3; ATP-binding cassette, sub-fa... 43.5 3e-04
mmu:192663 Abcg4, 6430517O04Rik; ATP-binding cassette, sub-fam... 43.5 3e-04
> eco:b4058 uvrA, dar, ECK4050, JW4019; ATPase and DNA damage
recognition protein of nucleotide excision repair excinuclease
UvrABC; K03701 excinuclease ABC subunit A
Length=940
Score = 246 bits (627), Expect = 3e-65, Method: Compositional matrix adjust.
Identities = 115/159 (72%), Positives = 136/159 (85%), Gaps = 0/159 (0%)
Query 3 EACHGDGIIKIEMHFLPDVYVPCEVCKGARYNRETLEVKYKGKTISDVLNMTVDEGCVFF 62
EAC GDG+IK+EMHFLPD+YVPC+ CKG RYNRETLE+KYKGKTI +VL+MT++E FF
Sbjct 741 EACQGDGVIKVEMHFLPDIYVPCDQCKGKRYNRETLEIKYKGKTIHEVLDMTIEEAREFF 800
Query 63 ENIPKIYRYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPT 122
+ +P + R ++ L DV L YI+LGQ ATTLSGGEAQRVKLA EL++R TG+TLYILDEPT
Sbjct 801 DAVPALARKLQTLMDVGLTYIRLGQSATTLSGGEAQRVKLARELSKRGTGQTLYILDEPT 860
Query 123 TGLHTADIHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
TGLH ADI QLLDVL +L D G+T+VVIEHNLDVIKTAD
Sbjct 861 TGLHFADIQQLLDVLHKLRDQGNTIVVIEHNLDVIKTAD 899
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 57/152 (37%), Positives = 83/152 (54%), Gaps = 15/152 (9%)
Query 24 PCEVCKGARYNRETLEVKYKGKTISDVLNMTVDEGCVFFENIP--------------KIY 69
PC C+G R RE V + + + +M++ FF N+ +I
Sbjct 405 PCASCEGTRLRREARHVYVENTPLPAISDMSIGHAMEFFNNLKLAGQRAKIAEKILKEIG 464
Query 70 RYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTAD 129
+K L +V L Y+ L + A TLSGGEAQR++LAS++ G +Y+LDEP+ GLH D
Sbjct 465 DRLKFLVNVGLNYLTLSRSAETLSGGEAQRIRLASQIGAGLVG-VMYVLDEPSIGLHQRD 523
Query 130 IHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
+LL L L D G+TV+V+EH+ D I+ AD
Sbjct 524 NERLLGTLIHLRDLGNTVIVVEHDEDAIRAAD 555
> dre:100003744 abcb10, si:dkey-162b3.3; ATP-binding cassette,
sub-family B (MDR/TAP), member 10; K05657 ATP-binding cassette,
subfamily B (MDR/TAP), member 10
Length=621
Score = 51.6 bits (122), Expect = 1e-06, Method: Composition-based stats.
Identities = 34/104 (32%), Positives = 56/104 (53%), Gaps = 11/104 (10%)
Query 65 IPKIYRYMKV------LQDVXLGY-IQLGQPATTLSGGEAQRVKLASELARRSTGKTLYI 117
+ IYR ++ +Q+ GY +G+ LSGG+ QRV +A L + + +
Sbjct 475 VEDIYRAAQIANAHDFIQEFPKGYDTVVGEKGVLLSGGQKQRVAIARALLKNPK---ILL 531
Query 118 LDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
LDE T+ L + + + L+RL+D G TV++I H L I++AD
Sbjct 532 LDEATSALDAENEFLVQEALERLMD-GRTVLIIAHRLSTIQSAD 574
> ath:AT1G31770 ABC transporter family protein
Length=648
Score = 50.8 bits (120), Expect = 2e-06, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 37/61 (60%), Gaps = 3/61 (4%)
Query 92 LSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIE 151
+SGGE +RV + E+ +L +LDEPT+GL + H+++ ++RL GG TVV
Sbjct 204 ISGGEKKRVSIGQEML---INPSLLLLDEPTSGLDSTTAHRIVTTIKRLASGGRTVVTTI 260
Query 152 H 152
H
Sbjct 261 H 261
> eco:b3454 livF, ECK3438, JW3419; leucine/isoleucine/valine transporter
subunit; K01996 branched-chain amino acid transport
system ATP-binding protein
Length=237
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 40/142 (28%), Positives = 66/142 (46%), Gaps = 22/142 (15%)
Query 31 ARYNRETLEVKYKGKTISDVLNMTVDEGCV----------FFENIPKIYRYMKVLQDVXL 80
A+ RE + + +G+ + MTV+E F E I +Y L + +
Sbjct 75 AKIMREAVAIVPEGRRVFS--RMTVEENLAMGGFFAERDQFQERIKWVYELFPRLHERRI 132
Query 81 GYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRL 140
Q A T+SGGE Q + + L + L +LDEP+ GL I Q+ D +++L
Sbjct 133 ------QRAGTMSGGEQQMLAIGRALM---SNPRLLLLDEPSLGLAPIIIQQIFDTIEQL 183
Query 141 VDGGDTVVVIEHNLD-VIKTAD 161
+ G T+ ++E N + +K AD
Sbjct 184 REQGMTIFLVEQNANQALKLAD 205
> hsa:23456 ABCB10, EST20237, M-ABC2, MTABC2; ATP-binding cassette,
sub-family B (MDR/TAP), member 10; K05657 ATP-binding
cassette, subfamily B (MDR/TAP), member 10
Length=738
Score = 49.3 bits (116), Expect = 6e-06, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 44/77 (57%), Gaps = 4/77 (5%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G+ LSGG+ QR+ +A L + + +LDE T+ L + + + + L RL+D G
Sbjct 627 VGEKGVLLSGGQKQRIAIARALLKNPK---ILLLDEATSALDAENEYLVQEALDRLMD-G 682
Query 145 DTVVVIEHNLDVIKTAD 161
TV+VI H L IK A+
Sbjct 683 RTVLVIAHRLSTIKNAN 699
> mmu:56199 Abcb10, AU016331, Abc-me, Abcb12, C76534; ATP-binding
cassette, sub-family B (MDR/TAP), member 10; K05657 ATP-binding
cassette, subfamily B (MDR/TAP), member 10
Length=715
Score = 48.9 bits (115), Expect = 7e-06, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 44/77 (57%), Gaps = 4/77 (5%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G+ LSGG+ QR+ +A L + + +LDE T+ L + H + + L RL++ G
Sbjct 592 VGEKGILLSGGQKQRIAIARALLKNP---KILLLDEATSALDAENEHLVQEALDRLME-G 647
Query 145 DTVVVIEHNLDVIKTAD 161
TV++I H L IK A+
Sbjct 648 RTVLIIAHRLSTIKNAN 664
> cel:W04C9.1 haf-4; HAlF transporter (PGP related) family member
(haf-4)
Length=787
Score = 48.9 bits (115), Expect = 8e-06, Method: Composition-based stats.
Identities = 25/77 (32%), Positives = 44/77 (57%), Gaps = 4/77 (5%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G+ T +SGG+ QR+ +A L R + +LDE T+ L T H + + + + +D G
Sbjct 629 VGEKGTQMSGGQKQRIAIARALVREP---AILLLDEATSALDTESEHLVQEAIYKNLD-G 684
Query 145 DTVVVIEHNLDVIKTAD 161
+V++I H L ++ AD
Sbjct 685 KSVILIAHRLSTVEKAD 701
> cel:ZK484.2 haf-9; HAlF transporter (PGP related) family member
(haf-9); K05656 ATP-binding cassette, subfamily B (MDR/TAP),
member 9
Length=815
Score = 48.5 bits (114), Expect = 9e-06, Method: Composition-based stats.
Identities = 26/89 (29%), Positives = 48/89 (53%), Gaps = 5/89 (5%)
Query 74 VLQDVXLGY-IQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQ 132
++ D GY +G+ + +SGG+ QR+ +A L R+ + +LDE T+ L H
Sbjct 643 IMNDTTDGYNTNVGEKGSQMSGGQKQRIAIARALVRQP---VVLLLDEATSALDAESEHT 699
Query 133 LLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
+ + + + + G TV++I H L ++ AD
Sbjct 700 VQEAISKNLK-GKTVILIAHRLSTVENAD 727
> mmu:74610 Abcb8, 4833412N02Rik, AA409895; ATP-binding cassette,
sub-family B (MDR/TAP), member 8; K05655 ATP-binding cassette,
subfamily B (MDR/TAP), member 8
Length=717
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 28/76 (36%), Positives = 42/76 (55%), Gaps = 4/76 (5%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G+ TTLSGG+ QR+ +A L ++ T+ ILDE T+ L + + L R G
Sbjct 587 VGERGTTLSGGQKQRLAIARALIKQP---TVLILDEATSALDAESERVVQEALDR-ASAG 642
Query 145 DTVVVIEHNLDVIKTA 160
TV+VI H L ++ A
Sbjct 643 RTVLVIAHRLSTVRAA 658
> ath:AT4G27420 ABC transporter family protein
Length=639
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 36/114 (31%), Positives = 55/114 (48%), Gaps = 16/114 (14%)
Query 52 NMTVDEGCVF---------FENIPKIYRYMKVLQDVXLGYIQ---LGQP-ATTLSGGEAQ 98
N+TV E VF F+ KI + V+ ++ L + +G P +SGGE +
Sbjct 140 NLTVTETLVFTALLRLPNSFKKQEKIKQAKAVMTELGLDRCKDTIIGGPFLRGVSGGERK 199
Query 99 RVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIEH 152
RV + E+ +L LDEPT+GL + +++ +L L GG TVV H
Sbjct 200 RVSIGQEIL---INPSLLFLDEPTSGLDSTTAQRIVSILWELARGGRTVVTTIH 250
> tgo:TGME49_060310 ATP-binding cassette protein subfamily B member
1 (EC:3.6.3.44); K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1345
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query 86 GQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDG-G 144
G + LSGG+ QRV +A L R ++ I DE T+ L A + L RL++ G
Sbjct 525 GSRGSQLSGGQKQRVAIARALIRHP---SILIFDEATSALDNASERVVQAALDRLIESTG 581
Query 145 DTVVVIEHNLDVIKTAD 161
T ++I H L I+ AD
Sbjct 582 VTTIIIAHRLSTIRRAD 598
Score = 33.5 bits (75), Expect = 0.31, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 33/66 (50%), Gaps = 5/66 (7%)
Query 98 QRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG--DTVVVIEHNLD 155
QR+ +A L R+ L ILDE T+ L Q+ L L++ G + +++ H L
Sbjct 1240 QRIAIARALVRKP---RLLILDEATSALDPESERQVQKTLDELMEKGHKHSTIIVAHRLS 1296
Query 156 VIKTAD 161
++ A+
Sbjct 1297 TVRNAN 1302
> hsa:11194 ABCB8, EST328128, M-ABC1, MABC1; ATP-binding cassette,
sub-family B (MDR/TAP), member 8; K05655 ATP-binding cassette,
subfamily B (MDR/TAP), member 8
Length=718
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 28/76 (36%), Positives = 42/76 (55%), Gaps = 4/76 (5%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G+ TTLSGG+ QR+ +A L ++ T+ ILDE T+ L + + L R G
Sbjct 588 VGERGTTLSGGQKQRLAIARALIKQP---TVLILDEATSALDAESERVVQEALDR-ASAG 643
Query 145 DTVVVIEHNLDVIKTA 160
TV+VI H L ++ A
Sbjct 644 RTVLVIAHRLSTVRGA 659
> hsa:6833 ABCC8, ABC36, HHF1, HI, HRINS, MRP8, PHHI, SUR, SUR1,
TNDM2; ATP-binding cassette, sub-family C (CFTR/MRP), member
8; K05032 ATP-binding cassette subfamily C (CFTR/MRP) member
8
Length=1581
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 37/124 (29%), Positives = 59/124 (47%), Gaps = 16/124 (12%)
Query 50 VLNMTVDEGCVFFENIPKIYRYMKVLQDVXL----------GYIQLGQPATTLSGGEAQR 99
+LN TV+E + FE+ RY V++ L Q+G+ LSGG+ QR
Sbjct 778 LLNATVEEN-IIFESPFNKQRYKMVIEACSLQPDIDILPHGDQTQIGERGINLSGGQRQR 836
Query 100 VKLASELARRSTGKTLYILDEPTTGL--HTADIHQLLDVLQRLVDGGDTVVVIEHNLDVI 157
+ +A L + + + LD+P + L H +D +L+ L D TVV++ H L +
Sbjct 837 ISVARALYQHA---NVVFLDDPFSALDIHLSDHLMQAGILELLRDDKRTVVLVTHKLQYL 893
Query 158 KTAD 161
AD
Sbjct 894 PHAD 897
> dre:100331842 hypothetical protein LOC100331842
Length=487
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 38/65 (58%), Gaps = 3/65 (4%)
Query 90 TTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVV 149
+ LSGG+ QRV +A LA G L + DEPT+ L + ++L L+ L + G T+V+
Sbjct 363 SELSGGQKQRVAIARALA---MGPELMLFDEPTSALDPEIVKEVLVFLRELAESGMTMVI 419
Query 150 IEHNL 154
+ H +
Sbjct 420 VTHEI 424
> dre:570453 abcg2b; ATP-binding cassette, sub-family G (WHITE),
member 2b
Length=618
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 35/61 (57%), Gaps = 3/61 (4%)
Query 92 LSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIE 151
+SGGE +R + EL T TL LDEPTTGL + + ++ +L RL GG T++
Sbjct 178 VSGGERKRCSIGMELI---TSPTLLFLDEPTTGLDSNTANSIIALLHRLSRGGRTIIFSI 234
Query 152 H 152
H
Sbjct 235 H 235
> dre:100334243 ATP-binding cassette, sub-family G (WHITE), member
2b-like
Length=618
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 35/61 (57%), Gaps = 3/61 (4%)
Query 92 LSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIE 151
+SGGE +R + EL T TL LDEPTTGL + + ++ +L RL GG T++
Sbjct 178 VSGGERKRCSIGMELI---TSPTLLFLDEPTTGLDSNTANSIIALLHRLSRGGRTIIFSI 234
Query 152 H 152
H
Sbjct 235 H 235
> tgo:TGME49_089350 white protein, putative (EC:3.6.3.28)
Length=766
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 43/76 (56%), Gaps = 4/76 (5%)
Query 77 DVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDV 136
D +G +Q+G +SGGE +R+ +A+E+ T ++ DEPTTGL + ++ V
Sbjct 271 DTCIGNLQMGA-RRGISGGEKKRLSVATEIV---TNPSIIFADEPTTGLDSFMAEAVMTV 326
Query 137 LQRLVDGGDTVVVIEH 152
L+RL G +++ H
Sbjct 327 LERLAQNGRSIICTIH 342
> dre:548340 abcb8, zgc:113037; ATP-binding cassette, sub-family
B (MDR/TAP), member 8; K05655 ATP-binding cassette, subfamily
B (MDR/TAP), member 8
Length=714
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 4/77 (5%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G+ TLSGG+ QR+ +A L + ++ ILDE T+ L + + L R G
Sbjct 603 VGERGVTLSGGQKQRIAIARALVKNP---SILILDEATSALDAESERVVQEALDRATT-G 658
Query 145 DTVVVIEHNLDVIKTAD 161
TV++I H L I+ AD
Sbjct 659 RTVLIIAHRLSTIQAAD 675
> eco:b2306 hisP, ECK2300, JW2303; histidine/lysine/arginine/ornithine
transporter subunit; K10017 histidine transport system
ATP-binding protein [EC:3.6.3.21]
Length=257
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 46/85 (54%), Gaps = 3/85 (3%)
Query 70 RYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTAD 129
R +K L V + G+ LSGG+ QRV +A LA + + DEPT+ L
Sbjct 131 RAVKYLAKVGIDERAQGKYPVHLSGGQQQRVSIARALAMEPE---VLLFDEPTSALDPEL 187
Query 130 IHQLLDVLQRLVDGGDTVVVIEHNL 154
+ ++L ++Q+L + G T+VV+ H +
Sbjct 188 VGEVLRIMQQLAEEGKTMVVVTHEM 212
> cel:F57A10.3 haf-3; HAlF transporter (PGP related) family member
(haf-3); K02021 putative ABC transport system ATP-binding
protein
Length=666
Score = 46.6 bits (109), Expect = 4e-05, Method: Composition-based stats.
Identities = 26/79 (32%), Positives = 45/79 (56%), Gaps = 4/79 (5%)
Query 84 QLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLV-D 142
++G+ +TLSGG+ QR+ +A L + + I+DE T+ L + + + L +L+ +
Sbjct 547 KVGEHGSTLSGGQKQRIAIARALVNKP---NILIMDEATSALDASSEYLVRIALNKLLAN 603
Query 143 GGDTVVVIEHNLDVIKTAD 161
TV++I H L IK AD
Sbjct 604 SKQTVMIIAHRLSTIKHAD 622
> eco:b0809 glnQ, ECK0798, JW0794; glutamine transporter subunit;
K10038 glutamine transport system ATP-binding protein [EC:3.6.3.-]
Length=240
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 39/65 (60%), Gaps = 3/65 (4%)
Query 90 TTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVV 149
+ LSGG+ QRV +A LA + + + DEPT+ L H++L V+Q L + G T+V+
Sbjct 135 SELSGGQQQRVAIARALAVKPK---MMLFDEPTSALDPELRHEVLKVMQDLAEEGMTMVI 191
Query 150 IEHNL 154
+ H +
Sbjct 192 VTHEI 196
> hsa:5243 ABCB1, ABC20, CD243, CLCS, GP170, MDR1, MGC163296,
P-GP, PGY1; ATP-binding cassette, sub-family B (MDR/TAP), member
1 (EC:3.6.3.44); K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1280
Score = 46.6 bits (109), Expect = 4e-05, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 48/101 (47%), Gaps = 16/101 (15%)
Query 61 FFENIPKIYRYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDE 120
F E++P Y ++G T LSGG+ QR+ +A L R+ + +LDE
Sbjct 1157 FIESLPNKYS------------TKVGDKGTQLSGGQKQRIAIARALVRQP---HILLLDE 1201
Query 121 PTTGLHTADIHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
T+ L T + + L + + G T +VI H L I+ AD
Sbjct 1202 ATSALDTESEKVVQEALDKARE-GRTCIVIAHRLSTIQNAD 1241
Score = 42.7 bits (99), Expect = 5e-04, Method: Composition-based stats.
Identities = 27/80 (33%), Positives = 41/80 (51%), Gaps = 10/80 (12%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHT---ADIHQLLDVLQRLV 141
+G+ LSGG+ QR+ +A L R + +LDE T+ L T A + LD ++
Sbjct 524 VGERGAQLSGGQKQRIAIARALVRNP---KILLLDEATSALDTESEAVVQVALDKARK-- 578
Query 142 DGGDTVVVIEHNLDVIKTAD 161
G T +VI H L ++ AD
Sbjct 579 --GRTTIVIAHRLSTVRNAD 596
> mmu:20927 Abcc8, D930031B21Rik, SUR1, Sur; ATP-binding cassette,
sub-family C (CFTR/MRP), member 8; K05032 ATP-binding cassette
subfamily C (CFTR/MRP) member 8
Length=1588
Score = 46.6 bits (109), Expect = 4e-05, Method: Composition-based stats.
Identities = 37/124 (29%), Positives = 59/124 (47%), Gaps = 16/124 (12%)
Query 50 VLNMTVDEGCVFFENIPKIYRYMKVLQDVXL----------GYIQLGQPATTLSGGEAQR 99
+LN TV+E + FE+ RY V++ L Q+G+ LSGG+ QR
Sbjct 785 LLNATVEEN-ITFESPFNKQRYKMVIEACSLQPDIDILPHGDQTQIGERGINLSGGQRQR 843
Query 100 VKLASELARRSTGKTLYILDEPTTGL--HTADIHQLLDVLQRLVDGGDTVVVIEHNLDVI 157
+ +A L + + + LD+P + L H +D +L+ L D TVV++ H L +
Sbjct 844 ISVARALYQHT---NVVFLDDPFSALDVHLSDHLMQAGILELLRDDKRTVVLVTHKLQYL 900
Query 158 KTAD 161
AD
Sbjct 901 PHAD 904
> ath:AT1G28010 PGP14; PGP14 (P-GLYCOPROTEIN 14); ATPase, coupled
to transmembrane movement of substances
Length=1247
Score = 46.2 bits (108), Expect = 4e-05, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 46/82 (56%), Gaps = 5/82 (6%)
Query 81 GYI-QLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQR 139
GY+ +G LSGG+ QRV +A + + ++ +LDE T+ L T+ Q+ + L +
Sbjct 1133 GYMTHVGDKGVQLSGGQKQRVAIARAVLK---DPSVLLLDEATSALDTSAEKQVQEALDK 1189
Query 140 LVDGGDTVVVIEHNLDVIKTAD 161
L+ G T +++ H L I+ AD
Sbjct 1190 LMK-GRTTILVAHRLSTIRKAD 1210
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 44/86 (51%), Gaps = 13/86 (15%)
Query 81 GY-IQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTAD---IHQLLD- 135
GY Q+G+ T LSGG+ QR+ +A + R + +LDE T+ L + Q LD
Sbjct 499 GYNTQVGEGGTQLSGGQKQRIAIARAVLRNP---KILLLDEATSALDAESEKIVQQALDN 555
Query 136 VLQRLVDGGDTVVVIEHNLDVIKTAD 161
V+++ T +VI H L I+ D
Sbjct 556 VMEK-----RTTIVIAHRLSTIRNVD 576
> mmu:18670 Abcb4, Mdr2, Pgy-2, Pgy2, mdr-2; ATP-binding cassette,
sub-family B (MDR/TAP), member 4 (EC:3.6.3.44); K05659
ATP-binding cassette, subfamily B (MDR/TAP), member 4
Length=1276
Score = 46.2 bits (108), Expect = 4e-05, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 48/101 (47%), Gaps = 16/101 (15%)
Query 61 FFENIPKIYRYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDE 120
F E +P+ Y ++G T LSGG+ QR+ +A L R+ + +LDE
Sbjct 1153 FIETLPQKYN------------TRVGDKGTQLSGGQKQRIAIARALIRQP---RVLLLDE 1197
Query 121 PTTGLHTADIHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
T+ L T + + L + + G T +VI H L I+ AD
Sbjct 1198 ATSALDTESEKVVQEALDKARE-GRTCIVIAHRLSTIQNAD 1237
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 28/80 (35%), Positives = 40/80 (50%), Gaps = 10/80 (12%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHT---ADIHQLLDVLQRLV 141
+G LSGG+ QR+ +A L R + +LDE T+ L T A++ LD +
Sbjct 523 VGDRGAQLSGGQKQRIAIARALVRNP---KILLLDEATSALDTESEAEVQAALDKARE-- 577
Query 142 DGGDTVVVIEHNLDVIKTAD 161
G T +VI H L I+ AD
Sbjct 578 --GRTTIVIAHRLSTIRNAD 595
> eco:b0879 macB, ECK0870, JW0863, ybjZ; fused macrolide transporter
subunits of ABC superfamily: ATP-binding component/membrane
component; K05685 macrolide transport system ATP-binding/permease
protein [EC:3.6.3.-]
Length=648
Score = 46.2 bits (108), Expect = 5e-05, Method: Composition-based stats.
Identities = 31/100 (31%), Positives = 51/100 (51%), Gaps = 4/100 (4%)
Query 62 FENIPKIYRYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEP 121
E ++ R ++LQ + L PA LSGG+ QRV +A L G + + DEP
Sbjct 116 LERKQRLLRAQELLQRLGLEDRTEYYPAQ-LSGGQQQRVSIARALM---NGGQVILADEP 171
Query 122 TTGLHTADIHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
T L + +++ +L +L D G TV+++ H+ V A+
Sbjct 172 TGALDSHSGEEVMAILHQLRDRGHTVIIVTHDPQVAAQAE 211
> ath:AT5G39040 ATTAP2; ATTAP2; ATPase, coupled to transmembrane
movement of substances / transporter
Length=644
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 43/77 (55%), Gaps = 4/77 (5%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G+ LSGG+ QR+ +A L T ++ +LDE T+ L + + D + L+ G
Sbjct 529 VGERGLRLSGGQKQRIAIARALL---TNPSVLLLDEATSALDAESEYLVQDAMDSLM-AG 584
Query 145 DTVVVIEHNLDVIKTAD 161
TV+VI H L +KTAD
Sbjct 585 RTVLVIAHRLSTVKTAD 601
> hsa:5244 ABCB4, ABC21, GBD1, MDR2, MDR2/3, MDR3, PFIC-3, PGY3;
ATP-binding cassette, sub-family B (MDR/TAP), member 4 (EC:3.6.3.44);
K05659 ATP-binding cassette, subfamily B (MDR/TAP),
member 4
Length=1279
Score = 46.2 bits (108), Expect = 5e-05, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 47/101 (46%), Gaps = 16/101 (15%)
Query 61 FFENIPKIYRYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDE 120
F E +P Y ++G T LSGG+ QR+ +A L R+ + +LDE
Sbjct 1156 FIETLPHKYE------------TRVGDKGTQLSGGQKQRIAIARALIRQP---QILLLDE 1200
Query 121 PTTGLHTADIHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
T+ L T + + L + + G T +VI H L I+ AD
Sbjct 1201 ATSALDTESEKVVQEALDKARE-GRTCIVIAHRLSTIQNAD 1240
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 27/80 (33%), Positives = 41/80 (51%), Gaps = 10/80 (12%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHT---ADIHQLLDVLQRLV 141
+G+ LSGG+ QR+ +A L R + +LDE T+ L T A++ LD +
Sbjct 526 VGERGAQLSGGQKQRIAIARALVRNP---KILLLDEATSALDTESEAEVQAALDKARE-- 580
Query 142 DGGDTVVVIEHNLDVIKTAD 161
G T +VI H L ++ AD
Sbjct 581 --GRTTIVIAHRLSTVRNAD 598
> mmu:18671 Abcb1a, Abcb4, Evi32, Mdr1a, Mdr3, P-gp, Pgp, Pgy-3,
Pgy3, mdr-3; ATP-binding cassette, sub-family B (MDR/TAP),
member 1A (EC:3.6.3.44); K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1276
Score = 45.8 bits (107), Expect = 6e-05, Method: Composition-based stats.
Identities = 27/78 (34%), Positives = 42/78 (53%), Gaps = 4/78 (5%)
Query 84 QLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDG 143
++G T LSGG+ QR+ +A L R+ + +LDE T+ L T + + L + +
Sbjct 1164 RVGDKGTQLSGGQKQRIAIARALVRQP---HILLLDEATSALDTESEKVVQEALDKARE- 1219
Query 144 GDTVVVIEHNLDVIKTAD 161
G T +VI H L I+ AD
Sbjct 1220 GRTCIVIAHRLSTIQNAD 1237
Score = 42.4 bits (98), Expect = 7e-04, Method: Composition-based stats.
Identities = 27/80 (33%), Positives = 40/80 (50%), Gaps = 10/80 (12%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHT---ADIHQLLDVLQRLV 141
+G+ LSGG+ QR+ +A L R + +LDE T+ L T A + LD +
Sbjct 520 VGERGAQLSGGQKQRIAIARALVRNP---KILLLDEATSALDTESEAVVQAALDKARE-- 574
Query 142 DGGDTVVVIEHNLDVIKTAD 161
G T +VI H L ++ AD
Sbjct 575 --GRTTIVIAHRLSTVRNAD 592
> dre:794238 abcg4b, zgc:171697; ATP-binding cassette, sub-family
G (WHITE), member 4b
Length=644
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 36/63 (57%), Gaps = 3/63 (4%)
Query 91 TLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVI 150
+LSGG+ +R+ +A EL + DEPT+GL +A Q++ +++ L GG T++
Sbjct 198 SLSGGQCKRLAIALELV---NNPPVMFFDEPTSGLDSASCFQVVSLMKSLAQGGRTIICT 254
Query 151 EHN 153
H
Sbjct 255 IHQ 257
> eco:b0652 gltL, ECK0645, JW0647; glutamate and aspartate transporter
subunit; K10004 glutamate/aspartate transport system
ATP-binding protein [EC:3.6.3.-]
Length=241
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 47/89 (52%), Gaps = 4/89 (4%)
Query 66 PKIYRYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGL 125
P + +K+L+ V L PA LSGG+ QRV +A L + DEPT+ L
Sbjct 112 PAREKALKLLERVGLSAHANKFPAQ-LSGGQQQRVAIARALCMDPIA---MLFDEPTSAL 167
Query 126 HTADIHQLLDVLQRLVDGGDTVVVIEHNL 154
I+++LDV+ L + G T++V+ H +
Sbjct 168 DPEMINEVLDVMVELANEGMTMMVVTHEM 196
> hsa:64241 ABCG8, GBD4, MGC142217, STSL; ATP-binding cassette,
sub-family G (WHITE), member 8; K05684 ATP-binding cassette,
subfamily G (WHITE), member 8 (sterolin 2)
Length=673
Score = 45.1 bits (105), Expect = 1e-04, Method: Composition-based stats.
Identities = 37/115 (32%), Positives = 55/115 (47%), Gaps = 18/115 (15%)
Query 52 NMTVDEGCVFFEN--IPKIYRYM---KVLQDVXLGYIQLGQPATT---------LSGGEA 97
N+TV E F +P+ + K ++DV + ++L Q A T LSGGE
Sbjct 160 NLTVRETLAFIAQMRLPRTFSQAQRDKRVEDV-IAELRLRQCADTRVGNMYVRGLSGGER 218
Query 98 QRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIEH 152
+RV + +L + ILDEPT+GL + H L+ L RL G V++ H
Sbjct 219 RRVSIGVQLL---WNPGILILDEPTSGLDSFTAHNLVKTLSRLAKGNRLVLISLH 270
> eco:b1858 znuC, ECK1859, JW1847, yebM; zinc transporter subunit:
ATP-binding component of ABC superfamily; K09817 zinc transport
system ATP-binding protein [EC:3.6.3.-]
Length=251
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 50/89 (56%), Gaps = 9/89 (10%)
Query 72 MKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHT---A 128
+ L+ V G++ + P LSGGE QRV LA L R L +LDEPT G+
Sbjct 102 LPALKRVQAGHL-INAPMQKLSGGETQRVLLARALLNRP---QLLVLDEPTQGVDVNGQV 157
Query 129 DIHQLLDVLQRLVDGGDTVVVIEHNLDVI 157
++ L+D L+R +D G V+++ H+L ++
Sbjct 158 ALYDLIDQLRRELDCG--VLMVSHDLHLV 184
> ath:AT1G02530 PGP12; PGP12 (P-GLYCOPROTEIN 12); ATPase, coupled
to transmembrane movement of substances
Length=1273
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 29/90 (32%), Positives = 44/90 (48%), Gaps = 5/90 (5%)
Query 73 KVLQDVXLGY-IQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIH 131
K + + GY +G+ LSGG+ QRV +A + + + +LDE T+ L
Sbjct 1149 KFISSIQQGYDTVVGEKGIQLSGGQKQRVAIARAIVKEPK---ILLLDEATSALDAESER 1205
Query 132 QLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
+ D L R++ T VV+ H L IK AD
Sbjct 1206 LVQDALDRVI-VNRTTVVVAHRLSTIKNAD 1234
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 42/78 (53%), Gaps = 4/78 (5%)
Query 84 QLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDG 143
++G+ T LSGG+ QR+ +A + + + +LDE T+ L T + + L R++
Sbjct 499 KVGEHGTQLSGGQKQRIAIARAILK---DPRVLLLDEATSALDTESERVVQEALDRVM-V 554
Query 144 GDTVVVIEHNLDVIKTAD 161
T VV+ H L ++ AD
Sbjct 555 NRTTVVVAHRLSTVRNAD 572
> dre:556979 abcg1, zgc:162197; ATP-binding cassette, sub-family
G (WHITE), member 1; K05679 ATP-binding cassette, subfamily
G (WHITE), member 1
Length=673
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 24/74 (32%), Positives = 42/74 (56%), Gaps = 4/74 (5%)
Query 80 LGYIQLGQPATT-LSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQ 138
LG ++ + T+ LSGG+ +R+ +A EL + DEPT+GL ++ Q++ +++
Sbjct 211 LGLLECAKTRTSHLSGGQRKRLAIALELV---NNPPVMFFDEPTSGLDSSSCFQVVSLMK 267
Query 139 RLVDGGDTVVVIEH 152
L GG TV+ H
Sbjct 268 ALAQGGRTVICTIH 281
> eco:b0914 msbA, ECK0905, JW0897; fused lipid transporter subunits
of ABC superfamily: membrane component/ATP-binding component;
K11085 ATP-binding cassette, subfamily B, bacterial
MsbA [EC:3.6.3.-]
Length=582
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 31/80 (38%), Positives = 41/80 (51%), Gaps = 10/80 (12%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTAD---IHQLLDVLQRLV 141
+G+ LSGG+ QR+ +A L R S + ILDE T+ L T I LD LQ+
Sbjct 474 IGENGVLLSGGQRQRIAIARALLRDS---PILILDEATSALDTESERAIQAALDELQK-- 528
Query 142 DGGDTVVVIEHNLDVIKTAD 161
T +VI H L I+ AD
Sbjct 529 --NRTSLVIAHRLSTIEKAD 546
> ath:AT2G01320 ABC transporter family protein
Length=725
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 34/61 (55%), Gaps = 3/61 (4%)
Query 92 LSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIE 151
+SGGE +R+ LA EL ++ DEPTTGL ++++ LQ+L G TV+
Sbjct 216 ISGGEKKRLSLACELI---ASPSVIFADEPTTGLDAFQAEKVMETLQKLAQDGHTVICSI 272
Query 152 H 152
H
Sbjct 273 H 273
> mmu:268379 Abca13, 9830132L24, A930002G16Rik, AI956815; ATP-binding
cassette, sub-family A (ABC1), member 13; K05647 ATP
binding cassette, subfamily A (ABC1), member 13
Length=5034
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 39/116 (33%), Positives = 60/116 (51%), Gaps = 13/116 (11%)
Query 49 DVL--NMTVDEGCVFFENIPKIYRYMKVLQ---DVXLGYIQLGQ----PATTLSGGEAQR 99
DVL N+TV E + F +I + K LQ + L ++L Q PA LSGG +
Sbjct 3892 DVLLDNLTVREHLMLFASIKAPWWTTKELQQQVNKTLDEVELTQHQHKPAGVLSGG--MK 3949
Query 100 VKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIEHNLD 155
KL+ +A KT+ +LDEP++G+ L D+L + + G T++ H+LD
Sbjct 3950 RKLSIGIAFMGMSKTV-VLDEPSSGVDPCSRRSLWDILLKYRE-GRTIIFTTHHLD 4003
Score = 32.0 bits (71), Expect = 1.0, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 36/70 (51%), Gaps = 5/70 (7%)
Query 87 QPATTLSGGEAQRVKLASELARRSTGK-TLYILDEPTTGLHTADIHQLLDVLQRLVDGGD 145
+P T SGG R KL++ LA GK + +LDEP++G+ L + + V G
Sbjct 4827 KPVATYSGG--TRRKLSTALAL--VGKPDILLLDEPSSGMDPCSKRYLWQTITQEVRDGC 4882
Query 146 TVVVIEHNLD 155
V+ H+++
Sbjct 4883 AAVLTSHSME 4892
> ath:AT3G28415 P-glycoprotein, putative
Length=1221
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 44/86 (51%), Gaps = 11/86 (12%)
Query 80 LGY-IQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQ 138
LGY Q+G+ +SGG+ QR+ +A + + TL +LDE T+ L + V+Q
Sbjct 464 LGYKTQVGERGVQMSGGQKQRISIARAIIK---SPTLLLLDEATSALDSESER----VVQ 516
Query 139 RLVDG---GDTVVVIEHNLDVIKTAD 161
+D G T +VI H L I+ D
Sbjct 517 EALDNATIGRTTIVIAHRLSTIRNVD 542
Score = 41.2 bits (95), Expect = 0.002, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 38/76 (50%), Gaps = 4/76 (5%)
Query 86 GQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGD 145
G LSGG+ QR+ +A + + + + +LDE T+ L + D L RL+ G
Sbjct 1111 GDRGVQLSGGQKQRIAIARAVLKNPS---VLLLDEATSALDNQSERMVQDALGRLM-VGR 1166
Query 146 TVVVIEHNLDVIKTAD 161
T VVI H L I+ D
Sbjct 1167 TSVVIAHRLSTIQNCD 1182
> mmu:67470 Abcg8, 1300003C16Rik, AI114946, sterolin-2; ATP-binding
cassette, sub-family G (WHITE), member 8; K05684 ATP-binding
cassette, subfamily G (WHITE), member 8 (sterolin 2)
Length=673
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 36/115 (31%), Positives = 55/115 (47%), Gaps = 18/115 (15%)
Query 52 NMTVDEGCVFFEN--IPKIYRYM---KVLQDVXLGYIQLGQPATT---------LSGGEA 97
N+TV E F +P+ + K ++DV + ++L Q A T +SGGE
Sbjct 161 NLTVRETLAFIAQMRLPRTFSQAQRDKRVEDV-IAELRLRQCANTRVGNTYVRGVSGGER 219
Query 98 QRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIEH 152
+RV + +L + ILDEPT+GL + H L+ L RL G V++ H
Sbjct 220 RRVSIGVQLL---WNPGILILDEPTSGLDSFTAHNLVTTLSRLAKGNRLVLISLH 271
> ath:AT4G25960 PGP2; PGP2 (P-GLYCOPROTEIN 2); ATPase, coupled
to transmembrane movement of substances
Length=1273
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 27/107 (25%), Positives = 50/107 (46%), Gaps = 16/107 (14%)
Query 55 VDEGCVFFENIPKIYRYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKT 114
+ E F N+P+ + Q+G+ LSGG+ QR+ ++ + + +
Sbjct 515 LSEAISFINNLPEGFE------------TQVGERGIQLSGGQKQRIAISRAIVKNPS--- 559
Query 115 LYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
+ +LDE T+ L + + L R++ G T VV+ H L ++ AD
Sbjct 560 ILLLDEATSALDAESEKSVQEALDRVM-VGRTTVVVAHRLSTVRNAD 605
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 26/82 (31%), Positives = 41/82 (50%), Gaps = 5/82 (6%)
Query 81 GY-IQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQR 139
GY ++G+ +SGG+ QR+ +A + + + +LDE T+ L + L R
Sbjct 1157 GYSTKVGERGVQMSGGQRQRIAIARAILKNPA---ILLLDEATSALDVESERVVQQALDR 1213
Query 140 LVDGGDTVVVIEHNLDVIKTAD 161
L+ T VV+ H L IK AD
Sbjct 1214 LM-ANRTTVVVAHRLSTIKNAD 1234
> tgo:TGME49_039020 ATP-binding cassette protein subfamily B member
2 (EC:3.6.3.30 3.6.3.44); K05658 ATP-binding cassette,
subfamily B (MDR/TAP), member 1
Length=1407
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 4/71 (5%)
Query 92 LSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGD-TVVVI 150
LSGG+ QR+ +A L RR ++ I DE T+ L T + D L L+ + T +++
Sbjct 585 LSGGQKQRIVIARALVRRP---SILIFDEATSALDTVSEKVVQDALDSLIKTTNATTLIV 641
Query 151 EHNLDVIKTAD 161
H L I+ AD
Sbjct 642 AHRLTTIRNAD 652
Score = 37.7 bits (86), Expect = 0.016, Method: Composition-based stats.
Identities = 26/83 (31%), Positives = 42/83 (50%), Gaps = 5/83 (6%)
Query 81 GY-IQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQR 139
GY Q+G+ + LSGG+ QR+ +A L T + ILDE T+ L + L
Sbjct 1285 GYDTQVGKGGSQLSGGQKQRIAIARALL---TQPRMLILDEATSALDAESERIVQATLDN 1341
Query 140 LVDGGDTV-VVIEHNLDVIKTAD 161
++ + V ++I H L ++ AD
Sbjct 1342 VIATKERVTLMIAHRLSTVRDAD 1364
> xla:443560 tap2, abc18, abcb3, apt2, psf2, ring11; transporter
2, ATP-binding cassette, sub-family B (MDR/TAP); K05654 ATP-binding
cassette, subfamily B (MDR/TAP), member 3
Length=714
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 5/77 (6%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G LS G+ QRV LA LAR+ L ILDE ++ L H++ LQ + D
Sbjct 601 VGDSGAQLSAGQKQRVALARALARKP---KLLILDEASSCLDAETEHEIQKSLQSIED-- 655
Query 145 DTVVVIEHNLDVIKTAD 161
++++I H L ++ AD
Sbjct 656 LSLLIIAHRLRTVQKAD 672
> hsa:340273 ABCB5, ABCB5alpha, ABCB5beta, EST422562; ATP-binding
cassette, sub-family B (MDR/TAP), member 5; K05660 ATP-binding
cassette, subfamily B (MDR/TAP), member 5
Length=1257
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G+ +SGG+ QR+ +A L R + ILDE T+ L + + L++ G
Sbjct 518 VGEKGAQMSGGQKQRIAIARALVRNP---KILILDEATSALDSESKSAVQAALEK-ASKG 573
Query 145 DTVVVIEHNLDVIKTAD 161
T +V+ H L I++AD
Sbjct 574 RTTIVVAHRLSTIRSAD 590
Score = 36.6 bits (83), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 48/104 (46%), Gaps = 22/104 (21%)
Query 61 FFENIPKIYRYMKVLQDVXLGYIQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDE 120
F E +P+ Y Q+G LSGG+ QR+ +A L ++ + +LDE
Sbjct 1137 FIEGLPEKYN------------TQVGLKGAQLSGGQKQRLAIARALLQKP---KILLLDE 1181
Query 121 PTTGLHTADIHQLLDVLQRLVDG---GDTVVVIEHNLDVIKTAD 161
T+ L D + V+Q +D G T +V+ H L I+ AD
Sbjct 1182 ATSALDN-DSEK---VVQHALDKARTGRTCLVVTHRLSAIQNAD 1221
> cel:Y48G8AL.11 haf-6; HAlF transporter (PGP related) family
member (haf-6); K05655 ATP-binding cassette, subfamily B (MDR/TAP),
member 8
Length=668
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 42/76 (55%), Gaps = 4/76 (5%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGG 144
+G+ LSGG+ QR+ +A + + + ILDE T+ L + H + + L ++ G
Sbjct 558 VGERGAQLSGGQKQRIAIARAILKNP---PILILDEATSALDSHSEHMVQEALNNVMK-G 613
Query 145 DTVVVIEHNLDVIKTA 160
TV++I H L I++A
Sbjct 614 RTVLIIAHRLSTIRSA 629
> xla:398090 TAP2 protein
Length=720
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 28/70 (40%), Positives = 39/70 (55%), Gaps = 5/70 (7%)
Query 92 LSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIE 151
LS G+ QRV LA LAR+ L ILDE ++ L H++ LQ L D ++++I
Sbjct 609 LSAGQKQRVALARALARKP---KLLILDEASSCLDAETEHEIQQSLQTLPD--VSLLIIA 663
Query 152 HNLDVIKTAD 161
H L I+ AD
Sbjct 664 HRLRTIQKAD 673
> cel:K08E7.9 pgp-1; P-GlycoProtein related family member (pgp-1)
Length=1321
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 41/78 (52%), Gaps = 4/78 (5%)
Query 84 QLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDG 143
++G T LSGG+ QR+ +A L R + +LDE T+ L T + + L R +
Sbjct 1210 RVGDRGTQLSGGQKQRIAIARALVRN---PKILLLDEATSALDTESEKVVQEALDRARE- 1265
Query 144 GDTVVVIEHNLDVIKTAD 161
G T +VI H L+ + AD
Sbjct 1266 GRTCIVIAHRLNTVMNAD 1283
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 47/96 (48%), Gaps = 14/96 (14%)
Query 70 RYMKVLQDVXLGYIQL-GQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTA 128
+++K L + GY L G T LSGG+ QR+ +A L R + +LDE T+ L
Sbjct 535 KFIKTLPN---GYNTLVGDRGTQLSGGQKQRIAIARALVRN---PKILLLDEATSALDAE 588
Query 129 D---IHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
+ Q LD + G T ++I H L I+ AD
Sbjct 589 SEGIVQQALDKAAK----GRTTIIIAHRLSTIRNAD 620
> cel:C34G6.4 pgp-2; P-GlycoProtein related family member (pgp-2)
Length=1272
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 43/81 (53%), Gaps = 10/81 (12%)
Query 84 QLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTAD---IHQLLDVLQRL 140
+G+ T LSGG+ QR+ +A L R ++ +LDE T+ L T + + LD ++
Sbjct 1160 HVGEKGTQLSGGQKQRIAIARALVR---SPSVLLLDEATSALDTESEKIVQEALDAAKQ- 1215
Query 141 VDGGDTVVVIEHNLDVIKTAD 161
G T +VI H L I+ +D
Sbjct 1216 ---GRTCLVIAHRLSTIQNSD 1233
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 46/92 (50%), Gaps = 8/92 (8%)
Query 71 YMKVLQDVXLGY-IQLGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTAD 129
++K L D GY ++G+ LSGG+ QR+ +A L + + +LDE T+ L T
Sbjct 519 FIKRLPD---GYGTRVGEKGVQLSGGQKQRIAIARALVKN---PKILLLDEATSALDTEA 572
Query 130 IHQLLDVLQRLVDGGDTVVVIEHNLDVIKTAD 161
++ L + G T +++ H L I+ D
Sbjct 573 EREVQGALDQ-AQAGRTTIIVAHRLSTIRNVD 603
> dre:570148 abcb9, si:dkey-21k4.3; ATP-binding cassette, sub-family
B (MDR/TAP), member 9; K05656 ATP-binding cassette, subfamily
B (MDR/TAP), member 9
Length=789
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 42/80 (52%), Gaps = 10/80 (12%)
Query 85 LGQPATTLSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTAD---IHQLLDVLQRLV 141
+G+ T LSGG+ QRV +A L R + ILDE T+ L + + Q L+ L R
Sbjct 643 VGEKGTQLSGGQKQRVAIARALIR---SPRILILDEATSALDSESEYIVQQALNNLMR-- 697
Query 142 DGGDTVVVIEHNLDVIKTAD 161
TV+VI H L ++ AD
Sbjct 698 --EHTVLVIAHRLSTVERAD 715
> mmu:192663 Abcg4, 6430517O04Rik; ATP-binding cassette, sub-family
G (WHITE), member 4; K05680 ATP-binding cassette, subfamily
G (WHITE), member 4
Length=646
Score = 43.5 bits (101), Expect = 3e-04, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 35/61 (57%), Gaps = 3/61 (4%)
Query 92 LSGGEAQRVKLASELARRSTGKTLYILDEPTTGLHTADIHQLLDVLQRLVDGGDTVVVIE 151
LSGG+ +R+ +A EL + DEPT+GL +A Q++ +++ L GG TV+
Sbjct 201 LSGGQRKRLAIALELV---NNPPVMFFDEPTSGLDSASCFQVVSLMKSLAHGGRTVICTI 257
Query 152 H 152
H
Sbjct 258 H 258
Lambda K H
0.320 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3702936828
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40