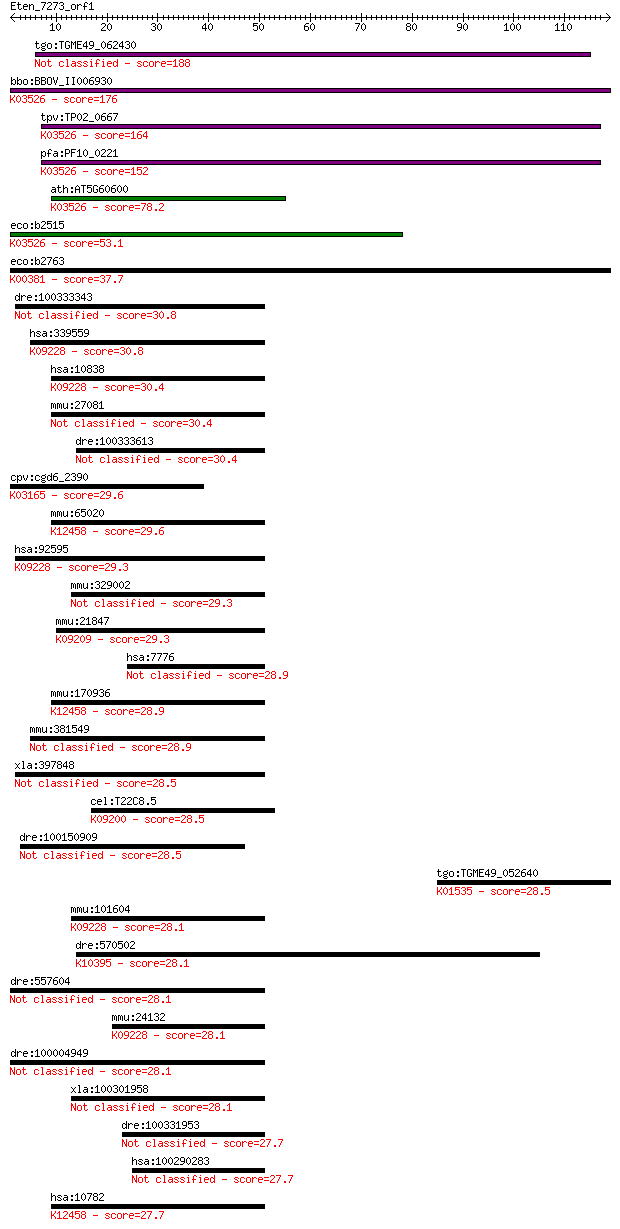
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7273_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_062430 4-hydroxy-3-methylbut-2-en-1-yl diphosphate ... 188 4e-48
bbo:BBOV_II006930 18.m06572; gcpE protein (EC:1.17.4.3); K0352... 176 1e-44
tpv:TP02_0667 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synt... 164 5e-41
pfa:PF10_0221 GcpE protein (EC:1.17.7.1); K03526 (E)-4-hydroxy... 152 2e-37
ath:AT5G60600 HDS; HDS (4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHA... 78.2 6e-15
eco:b2515 ispG, ECK2511, gcpE, JW2499; 1-hydroxy-2-methyl-2-(E... 53.1 2e-07
eco:b2763 cysI, ECK2758, JW2733; sulfite reductase, beta subun... 37.7 0.008
dre:100333343 zinc finger protein 45-like 30.8 1.0
hsa:339559 ZNF642, FLJ16030, RP11-656D10.2, Zfp69; zinc finger... 30.8 1.1
hsa:10838 ZNF275; zinc finger protein 275; K09228 KRAB domain-... 30.4 1.3
mmu:27081 Zfp275, 5430431L06Rik, AI593314, DXBay20, DXHXS52, D... 30.4 1.4
dre:100333613 zinc finger protein 300-like 30.4 1.5
cpv:cgd6_2390 DNA topoisomerase III beta-1 ; K03165 DNA topois... 29.6 2.3
mmu:65020 Zfp110, 2900024E01Rik, N28112, NRIF, Nrif1; zinc fin... 29.6 2.3
hsa:92595 ZNF764, MGC13138; zinc finger protein 764; K09228 KR... 29.3 2.9
mmu:329002 Zfp236, AI447957, Gm335; zinc finger protein 236 29.3
mmu:21847 Klf10, AI115143, EGR[a], Egral, Gdnfif, Tieg, Tieg1,... 29.3 3.5
hsa:7776 ZNF236, ZNF236A, ZNF236B; zinc finger protein 236 28.9
mmu:170936 Zfp369, B930030B22Rik, D230020H11Rik, NRIF2; zinc f... 28.9 4.4
mmu:381549 Zfp69, Gm1029, Krab2, Zfp63; zinc finger protein 69 28.9
xla:397848 prdm9, XFDL141; PR domain containing 9 (EC:2.1.1.43) 28.5 4.7
cel:T22C8.5 sptf-2; Specificity Protein) Transcription Factor ... 28.5 5.0
dre:100150909 si:ch211-241b2.4 28.5 5.5
tgo:TGME49_052640 plasma-membrane H+-ATPase, putative (EC:3.6.... 28.5 6.0
mmu:101604 E430018J23Rik, AI480612; RIKEN cDNA E430018J23 gene... 28.1 6.2
dre:570502 hCG2027369-like; K10395 kinesin family member 4/7/2... 28.1 6.6
dre:557604 zinc finger protein 551-like 28.1 6.8
mmu:24132 Zfp53, D030067O06Rik, KRAZ1, Zfp-53, Zfp118, zfas8; ... 28.1 7.1
dre:100004949 zinc finger protein 551-like 28.1 7.3
xla:100301958 hypothetical protein LOC100301958 28.1 8.0
dre:100331953 zinc finger protein 300-like 27.7 8.4
hsa:100290283 zinc finger protein 136-like 27.7 8.4
hsa:10782 ZNF274, DKFZp686K08243, FLJ37843, HFB101, ZF2, ZKSCA... 27.7 8.4
> tgo:TGME49_062430 4-hydroxy-3-methylbut-2-en-1-yl diphosphate
synthase, putative (EC:1.17.4.3)
Length=1283
Score = 188 bits (477), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 83/109 (76%), Positives = 98/109 (89%), Gaps = 0/109 (0%)
Query 6 GLGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLPGVKIAVMGCIVNGIG 65
G LLQACRLR++KTEFISCPSCGRTLFDIQ+V+AEI++ GHLPGV IAVMGCIVNG+G
Sbjct 1164 GFNLLQACRLRNTKTEFISCPSCGRTLFDIQKVSAEIRRRTGHLPGVTIAVMGCIVNGVG 1223
Query 66 EMGDADFGYVGGAPGLVDLYVGKRLARRGVKSAEACEALIELIKKEGRW 114
EM DADFGYVGGAPG VDLYV K++ +RG+ SAEAC+AL++LIK+ GRW
Sbjct 1224 EMADADFGYVGGAPGKVDLYVKKKVVKRGISSAEACDALVDLIKQHGRW 1272
> bbo:BBOV_II006930 18.m06572; gcpE protein (EC:1.17.4.3); K03526
(E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase [EC:1.17.7.1]
Length=772
Score = 176 bits (446), Expect = 1e-44, Method: Composition-based stats.
Identities = 82/118 (69%), Positives = 89/118 (75%), Gaps = 0/118 (0%)
Query 1 ERTQKGLGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLPGVKIAVMGCI 60
E L +LQA R+RS KTEFISCPSCGRTLFDIQ+ IKK +GHLPGV IA+MGCI
Sbjct 655 ESNDIALSILQAARMRSFKTEFISCPSCGRTLFDIQKTTESIKKRVGHLPGVSIAIMGCI 714
Query 61 VNGIGEMGDADFGYVGGAPGLVDLYVGKRLARRGVKSAEACEALIELIKKEGRWKEKE 118
VNGIGEM DADFGYVGGAP VDLYV K L R V EAC+ LIELIKK G+W E E
Sbjct 715 VNGIGEMADADFGYVGGAPHKVDLYVKKELVERNVPDVEACDRLIELIKKHGKWVEPE 772
> tpv:TP02_0667 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
K03526 (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate
synthase [EC:1.17.7.1]
Length=801
Score = 164 bits (416), Expect = 5e-41, Method: Composition-based stats.
Identities = 71/110 (64%), Positives = 88/110 (80%), Gaps = 0/110 (0%)
Query 7 LGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLPGVKIAVMGCIVNGIGE 66
L +LQ R+R+ KT+FISCPSCGRTLF+IQ+ E+++ +GHLPGV IA+MGCIVNGIGE
Sbjct 690 LDILQNSRMRNIKTDFISCPSCGRTLFNIQETTEEVRRRIGHLPGVTIAIMGCIVNGIGE 749
Query 67 MGDADFGYVGGAPGLVDLYVGKRLARRGVKSAEACEALIELIKKEGRWKE 116
M DADFGYVGG+PG VDLYV K+L R + +AC+ LIELIKK +W E
Sbjct 750 MADADFGYVGGSPGKVDLYVKKQLIERNIPKDQACDKLIELIKKHNKWVE 799
> pfa:PF10_0221 GcpE protein (EC:1.17.7.1); K03526 (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate
synthase [EC:1.17.7.1]
Length=824
Score = 152 bits (385), Expect = 2e-37, Method: Composition-based stats.
Identities = 69/110 (62%), Positives = 82/110 (74%), Gaps = 0/110 (0%)
Query 7 LGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLPGVKIAVMGCIVNGIGE 66
L +LQ R+R KT++I+CPSCGRTLF+IQ+ +I K GHL GVKIAVMGCIVNGIGE
Sbjct 714 LNILQDTRIRLFKTDYIACPSCGRTLFNIQETTKKIMKLTGHLKGVKIAVMGCIVNGIGE 773
Query 67 MGDADFGYVGGAPGLVDLYVGKRLARRGVKSAEACEALIELIKKEGRWKE 116
M DA FGYVG AP +DLY GK L R + EAC+ LIELIKK +WK+
Sbjct 774 MADAHFGYVGSAPKKIDLYYGKELVERNIPEEEACDKLIELIKKHNKWKD 823
> ath:AT5G60600 HDS; HDS (4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE
SYNTHASE); 4 iron, 4 sulfur cluster binding / 4-hydroxy-3-methylbut-2-en-1-yl
diphosphate synthase (EC:1.17.7.1); K03526
(E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase
[EC:1.17.7.1]
Length=717
Score = 78.2 bits (191), Expect = 6e-15, Method: Composition-based stats.
Identities = 29/46 (63%), Positives = 41/46 (89%), Gaps = 0/46 (0%)
Query 9 LLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLPGVKI 54
LLQ CR+R++KTE++SCPSCGRTLFD+Q+++AEI++ HLPGV +
Sbjct 628 LLQGCRMRNTKTEYVSCPSCGRTLFDLQEISAEIREKTSHLPGVSV 673
> eco:b2515 ispG, ECK2511, gcpE, JW2499; 1-hydroxy-2-methyl-2-(E)-butenyl
4-diphosphate synthase (EC:1.17.7.1); K03526 (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate
synthase [EC:1.17.7.1]
Length=372
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 44/78 (56%), Gaps = 1/78 (1%)
Query 1 ERTQKGLGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGH-LPGVKIAVMGC 59
E + G +L++ R+RS FI+CP+C R FD+ +++ L + + ++++GC
Sbjct 246 EEIKVGFDILKSLRIRSRGINFIACPTCSRQEFDVIGTVNALEQRLEDIITPMDVSIIGC 305
Query 60 IVNGIGEMGDADFGYVGG 77
+VNG GE + G GG
Sbjct 306 VVNGPGEALVSTLGVTGG 323
> eco:b2763 cysI, ECK2758, JW2733; sulfite reductase, beta subunit,
NAD(P)-binding, heme-binding (EC:1.8.1.2); K00381 sulfite
reductase (NADPH) hemoprotein beta-component [EC:1.8.1.2]
Length=570
Score = 37.7 bits (86), Expect = 0.008, Method: Composition-based stats.
Identities = 39/136 (28%), Positives = 59/136 (43%), Gaps = 22/136 (16%)
Query 1 ERTQKGLGLLQACR-LRSSKTEFISCPSCGRTLFD--------IQQVAAEIKKHLGHLPG 51
E+ K GL+ A R + +S P+C + + I + + KH
Sbjct 412 EKIAKESGLMNAVTPQRENSMACVSFPTCPLAMAEAERFLPSFIDNIDNLMAKHGVSDEH 471
Query 52 VKIAVMGCIVNGIGEMGDADFGYVGGAPGLVDLYVG--------KRLARRGVKSAEACEA 103
+ + V GC NG G A+ G VG APG +L++G R+ + + E +
Sbjct 472 IVMRVTGC-PNGCGRAMLAEVGLVGKAPGRYNLHLGGNRIGTRIPRMYKENITEPEILAS 530
Query 104 LIELIKKEGRW-KEKE 118
L ELI GRW KE+E
Sbjct 531 LDELI---GRWAKERE 543
> dre:100333343 zinc finger protein 45-like
Length=481
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 25/53 (47%), Gaps = 5/53 (9%)
Query 2 RTQKGLGLLQA----CRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
+ QK L QA R+ + + F SC CGR+ IQ + +K H G P
Sbjct 338 QCQKSFRLKQALEIHTRIHTGEKPF-SCQQCGRSFSQIQSIKHHMKTHTGEKP 389
> hsa:339559 ZNF642, FLJ16030, RP11-656D10.2, Zfp69; zinc finger
protein 642; K09228 KRAB domain-containing zinc finger protein
Length=526
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query 5 KGLGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
+ + L+Q R S + F +C CG+T I+ ++ I+ H G P
Sbjct 366 QNISLVQHLRTHSGEKPF-TCNECGKTFRQIRHLSEHIRIHTGEKP 410
> hsa:10838 ZNF275; zinc finger protein 275; K09228 KRAB domain-containing
zinc finger protein
Length=329
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 9 LLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
L + +L +S+ F +C +C R D Q++ + H GHLP
Sbjct 224 LFRHQKLHTSEKPF-ACKACSRDFLDRQELLKHQRMHTGHLP 264
> mmu:27081 Zfp275, 5430431L06Rik, AI593314, DXBay20, DXHXS52,
DXPas8; zinc finger protein 275
Length=377
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query 9 LLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
L + +L +S+ F +C +C R D Q++ + H GHLP
Sbjct 172 LFRHQKLHTSEKPF-ACKACSRDFLDRQELLKHQRMHTGHLP 212
> dre:100333613 zinc finger protein 300-like
Length=381
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query 14 RLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
R+ + + F SCP CG++ D Q ++ H G P
Sbjct 248 RIHTGEKPF-SCPQCGKSFIDKQNFKVHMRVHTGEKP 283
> cpv:cgd6_2390 DNA topoisomerase III beta-1 ; K03165 DNA topoisomerase
III [EC:5.99.1.2]
Length=835
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 12/39 (30%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 1 ERTQK-GLGLLQACRLRSSKTEFISCPSCGRTLFDIQQV 38
ER +K G L+ C+ ++S+ F+ CP+C + + +V
Sbjct 770 ERCEKCGSRKLKICKEKTSQEHFVGCPNCDEEILSLIEV 808
> mmu:65020 Zfp110, 2900024E01Rik, N28112, NRIF, Nrif1; zinc finger
protein 110; K12458 zinc finger protein 274
Length=832
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 20/42 (47%), Gaps = 1/42 (2%)
Query 9 LLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
L Q R+ S + F C CGRT D ++ ++ H G P
Sbjct 731 LTQHLRIHSGERPF-ECSECGRTFNDRSAISQHLRTHTGAKP 771
> hsa:92595 ZNF764, MGC13138; zinc finger protein 764; K09228
KRAB domain-containing zinc finger protein
Length=407
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 21/49 (42%), Gaps = 1/49 (2%)
Query 2 RTQKGLGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
R + L Q R+ S +T F CP CGR + ++ H G P
Sbjct 266 RFSQSSALYQHRRVHSGETPF-PCPDCGRAFAYPSDLRRHVRTHTGEKP 313
> mmu:329002 Zfp236, AI447957, Gm335; zinc finger protein 236
Length=1799
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 21/49 (42%), Gaps = 11/49 (22%)
Query 13 CRLRSSKTEFI-----------SCPSCGRTLFDIQQVAAEIKKHLGHLP 50
C++R+S T SCP CG+T Q+ I+ H G P
Sbjct 178 CKVRASGTRSYNRNIDRSSFPYSCPHCGKTFQKPSQLTRHIRIHTGERP 226
> mmu:21847 Klf10, AI115143, EGR[a], Egral, Gdnfif, Tieg, Tieg1,
mGIF; Kruppel-like factor 10; K09209 krueppel-like factor
10/11
Length=479
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 20/41 (48%), Gaps = 3/41 (7%)
Query 10 LQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
+ + R+RS S P CG+T F + A ++ H G P
Sbjct 360 IDSSRVRS---HICSHPGCGKTYFKSSHLKAHVRTHTGEKP 397
> hsa:7776 ZNF236, ZNF236A, ZNF236B; zinc finger protein 236
Length=1845
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 14/27 (51%), Gaps = 0/27 (0%)
Query 24 SCPSCGRTLFDIQQVAAEIKKHLGHLP 50
SCP CG+T Q+ I+ H G P
Sbjct 198 SCPHCGKTFQKPSQLTRHIRIHTGERP 224
> mmu:170936 Zfp369, B930030B22Rik, D230020H11Rik, NRIF2; zinc
finger protein 369; K12458 zinc finger protein 274
Length=845
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 19/42 (45%), Gaps = 1/42 (2%)
Query 9 LLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
L Q R+ S + F C CGRT D + ++ H G P
Sbjct 744 LTQHLRIHSGERPF-ECSECGRTFSDRSAASQHLRTHTGAKP 784
> mmu:381549 Zfp69, Gm1029, Krab2, Zfp63; zinc finger protein
69
Length=587
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 13/46 (28%), Positives = 23/46 (50%), Gaps = 1/46 (2%)
Query 5 KGLGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
+ + L+Q R S + F SC CG+ I+ ++ ++ H G P
Sbjct 419 QNISLVQHLRTHSGEKPF-SCSECGKPFRQIRHLSEHVRIHTGEKP 463
> xla:397848 prdm9, XFDL141; PR domain containing 9 (EC:2.1.1.43)
Length=411
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 25/49 (51%), Gaps = 3/49 (6%)
Query 2 RTQKGLGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
+T+ L L C + + + F+ CP CG+ D + + +++H G P
Sbjct 329 KTKASLAL--HCHIHTGERPFV-CPECGKGFRDPTSLNSHVRRHTGEKP 374
> cel:T22C8.5 sptf-2; Specificity Protein) Transcription Factor
family member (sptf-2); K09200 transcription factor Sp, invertebrate
Length=166
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 17/36 (47%), Gaps = 0/36 (0%)
Query 17 SSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLPGV 52
S T S P CG+T + A ++KH G P V
Sbjct 73 SQHTHLCSVPGCGKTYKKTSHLRAHLRKHTGDRPFV 108
> dre:100150909 si:ch211-241b2.4
Length=294
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Query 3 TQKGLGLLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHL 46
TQ G L+Q R+ + + +I C CGR+ + + A IKKHL
Sbjct 242 TQSG-HLMQHERVHTGRKPYI-CSICGRSFSTARNLPAHIKKHL 283
> tgo:TGME49_052640 plasma-membrane H+-ATPase, putative (EC:3.6.3.6);
K01535 H+-transporting ATPase [EC:3.6.3.6]
Length=1039
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 9/34 (26%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 85 YVGKRLARRGVKSAEACEALIELIKKEGRWKEKE 118
Y+G+R A + + E A + +K++G+W+ ++
Sbjct 109 YIGQRSAHNAIAAVEKLGAPVCQVKRDGQWQNRQ 142
> mmu:101604 E430018J23Rik, AI480612; RIKEN cDNA E430018J23 gene;
K09228 KRAB domain-containing zinc finger protein
Length=461
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 18/39 (46%), Gaps = 1/39 (2%)
Query 13 CRLRSSKTEF-ISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
C +R+ E SCP CGR +AA + H G P
Sbjct 354 CHMRTHTGEKPYSCPDCGRCFRQSSDMAAHRRTHSGERP 392
> dre:570502 hCG2027369-like; K10395 kinesin family member 4/7/21/27
Length=1651
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 41/103 (39%), Gaps = 12/103 (11%)
Query 14 RLRSSKTEFISCPSCG---RTLFDIQQVAAEIKKH---LGHLPGVKIAVMGCIVNG---- 63
RL SS+ E + C G RT Q A + H HL +++ +VNG
Sbjct 189 RLVSSEDELLQCLKLGALSRTTASTQMNAQSSRSHAIFTIHLCQMRVCPQPQLVNGELNG 248
Query 64 --IGEMGDADFGYVGGAPGLVDLYVGKRLARRGVKSAEACEAL 104
G M +++ + VDL +RL R G A E +
Sbjct 249 VSDGSMSQSEYETLSAKFHFVDLAGSERLKRTGATGERAREGI 291
> dre:557604 zinc finger protein 551-like
Length=420
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 14/54 (25%), Positives = 27/54 (50%), Gaps = 5/54 (9%)
Query 1 ERTQKGLGLLQA----CRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
E+ K G +Q R+ + + +F +C CG++ + + AA ++ H G P
Sbjct 113 EQCGKSFGQIQGFKAHMRIHTGERKF-TCQECGKSFYHVGNFAAHMRIHTGEKP 165
> mmu:24132 Zfp53, D030067O06Rik, KRAZ1, Zfp-53, Zfp118, zfas8;
zinc finger protein 53; K09228 KRAB domain-containing zinc
finger protein
Length=652
Score = 28.1 bits (61), Expect = 7.1, Method: Composition-based stats.
Identities = 10/30 (33%), Positives = 15/30 (50%), Gaps = 0/30 (0%)
Query 21 EFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
E SCP CG+ L + + + + H G P
Sbjct 336 EHCSCPECGKVLHQLSHLRSHYRLHTGEKP 365
> dre:100004949 zinc finger protein 551-like
Length=420
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 14/54 (25%), Positives = 27/54 (50%), Gaps = 5/54 (9%)
Query 1 ERTQKGLGLLQA----CRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
E+ K G +Q R+ + + +F +C CG++ + + AA ++ H G P
Sbjct 113 EQCGKSFGQIQGFKAHMRIHTGERKF-TCQECGKSFYHVGNFAAHMRIHTGEKP 165
> xla:100301958 hypothetical protein LOC100301958
Length=439
Score = 28.1 bits (61), Expect = 8.0, Method: Composition-based stats.
Identities = 10/38 (26%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Query 13 CRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
C + + + F+ CP CG+ D + + +++H G P
Sbjct 366 CHIHTGEKPFV-CPECGKGYRDPTSLNSHVRRHTGEEP 402
> dre:100331953 zinc finger protein 300-like
Length=301
Score = 27.7 bits (60), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 9/28 (32%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 23 ISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
SC CG++ IQ + ++ H G++P
Sbjct 160 FSCQQCGKSFTQIQNLNVHMRVHTGNMP 187
> hsa:100290283 zinc finger protein 136-like
Length=400
Score = 27.7 bits (60), Expect = 8.4, Method: Composition-based stats.
Identities = 9/26 (34%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 25 CPSCGRTLFDIQQVAAEIKKHLGHLP 50
C CG+T F ++++ I H G+ P
Sbjct 170 CKECGKTFFSLKRIRRHIITHSGYTP 195
> hsa:10782 ZNF274, DKFZp686K08243, FLJ37843, HFB101, ZF2, ZKSCAN19;
zinc finger protein 274; K12458 zinc finger protein 274
Length=548
Score = 27.7 bits (60), Expect = 8.4, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 20/42 (47%), Gaps = 1/42 (2%)
Query 9 LLQACRLRSSKTEFISCPSCGRTLFDIQQVAAEIKKHLGHLP 50
L Q R+ S + F C CGRT D ++ ++ H G P
Sbjct 445 LTQHQRVHSGERPF-ECQECGRTFNDRSAISQHLRTHTGAKP 485
Lambda K H
0.321 0.140 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40