bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7259_orf1
Length=157
Score E
Sequences producing significant alignments: (Bits) Value
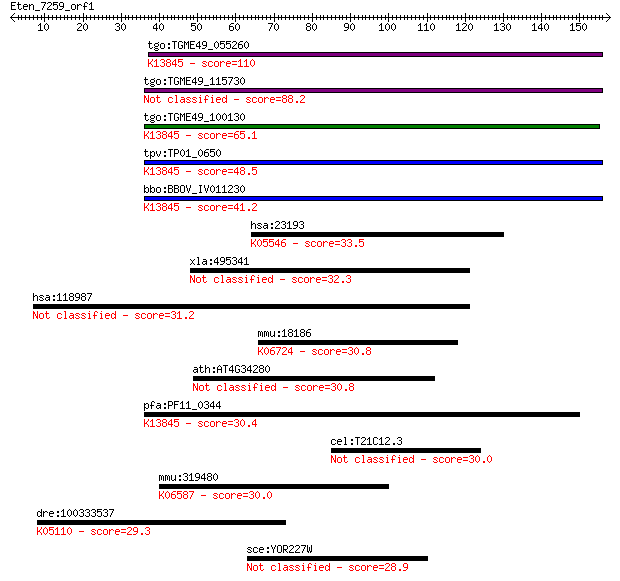
tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845... 110 2e-24
tgo:TGME49_115730 apical membrane antigen, putative 88.2 1e-17
tgo:TGME49_100130 apical membrane antigen, putative ; K13845 a... 65.1 1e-10
tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoi... 48.5 1e-05
bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845... 41.2 0.002
hsa:23193 GANAB, G2AN, GLUII, KIAA0088; glucosidase, alpha; ne... 33.5 0.29
xla:495341 mtmr10-a, mtmr10; myotubularin related protein 10 32.3
hsa:118987 PDZD8, FLJ25412, FLJ34427, PDZK8, bA129M16.2; PDZ d... 31.2 1.3
mmu:18186 Nrp1, C530029I03, NP-1, NPN-1, Npn1, Nrp; neuropilin... 30.8 2.0
ath:AT4G34280 transducin family protein / WD-40 repeat family ... 30.8 2.1
pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane ... 30.4 2.2
cel:T21C12.3 hypothetical protein 30.0 2.9
mmu:319480 Itga11, 4732459H24Rik; integrin alpha 11; K06587 in... 30.0 3.6
dre:100333537 ephb1; EPH receptor B1; K05110 Eph receptor B1 [... 29.3 5.7
sce:YOR227W HER1; Her1p 28.9 7.2
> tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845
apical merozoite antigen 1
Length=569
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 58/125 (46%), Positives = 78/125 (62%), Gaps = 7/125 (5%)
Query 37 CPVFGKNIEFYQPLDSDLYKNDFLENVPTE-EAAAAAKPLPGGFNNNFLMKDKKPFSPMS 95
CP++GK+IE QP D Y+N+FLE+VPTE E + PLPGGFN NF+ + SP
Sbjct 117 CPIWGKHIELQQP-DRPPYRNNFLEDVPTEKEYKQSGNPLPGGFNLNFVTPSGQRISPFP 175
Query 96 VAQLNSYPQLKARTGLGKCAEMSYLTTA-----AGSSYRYPFVFDSKKDLCYLLLVPLQR 150
+ L +KA T LG+CAE ++ T A + YRYPFV+DSKK LC++L V +Q
Sbjct 176 MELLEKNSNIKASTDLGRCAEFAFKTVAMDKNNKATKYRYPFVYDSKKRLCHILYVSMQL 235
Query 151 LMGGK 155
+ G K
Sbjct 236 MEGKK 240
> tgo:TGME49_115730 apical membrane antigen, putative
Length=388
Score = 88.2 bits (217), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 51/127 (40%), Positives = 72/127 (56%), Gaps = 11/127 (8%)
Query 36 RCPVFGKNIEFYQPLDSDLYKNDFLENVPTEEAAAAAKPLPGGFNNNFLMKDKKPFSPMS 95
+CPVFGK I+ +QP + Y N+FL++ PT A+ KPLPGGFNN + + FSP+
Sbjct 140 KCPVFGKAIQMHQPAE---YSNNFLDDAPTSNDASK-KPLPGGFNNPQVYTSGQKFSPID 195
Query 96 VAQLNS-YPQLKARTGLGKCAEMSYLTTAAG------SSYRYPFVFDSKKDLCYLLLVPL 148
+ L +T +G+CA +Y T A S+Y+YPFV+D+ CY+L V
Sbjct 196 DSLLQERLGTAGPKTAIGRCALYAYSTIAVNPSTNYTSTYKYPFVYDAVSRKCYVLSVSA 255
Query 149 QRLMGGK 155
Q L G K
Sbjct 256 QLLKGEK 262
> tgo:TGME49_100130 apical membrane antigen, putative ; K13845
apical merozoite antigen 1
Length=493
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 44/146 (30%), Positives = 68/146 (46%), Gaps = 32/146 (21%)
Query 36 RCPVFGKNIEFYQPL-DSDLYKNDFLENVPTEEAAAAAKPLPGGFNNNFLMKDKKPFSPM 94
+CP +GK I+ YQP + +++ NDFL+ VP PL GGF SP+
Sbjct 95 KCPNYGKYIKTYQPTTNPEIWPNDFLKPVPYANTPQDTMPLGGGF-----AMPMHQISPV 149
Query 95 SVAQLNSYPQ-LKARTG---------------LGKCAEMSYLTTAAGSS----------Y 128
S+ L + LK TG LG C + +T+A +S Y
Sbjct 150 SLKDLKDEAEGLKTATGVSSYAVEHAKNIRDDLGHCIWWARMTSAHDTSSSATNSKEDYY 209
Query 129 RYPFVFDSKKDLCYLLLVPLQRLMGG 154
RY FV+D KK++C+++ + +Q + G
Sbjct 210 RYAFVWDPKKEMCHIMYLNMQEMTGA 235
> tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoite
antigen 1
Length=785
Score = 48.5 bits (114), Expect = 1e-05, Method: Composition-based stats.
Identities = 39/128 (30%), Positives = 56/128 (43%), Gaps = 18/128 (14%)
Query 36 RCPVFGKNIEFYQPLDSDLYKN--DFLENVPTEEAAAAAKPLPG---GFNNNFLMKDKKP 90
+CPV GK I + +N DFL ++ + P N++ + +
Sbjct 288 KCPVVGKAI---------ILENGADFLSSITHHDPKERGLGFPATKVASNSSKQDMENQL 338
Query 91 FSPMSVAQLNSYPQLKARTGLGKCAEMSYLTTAAG---SSYRYPFVFDSKKDLCYLLLVP 147
SP+S L S+ K + L CAE S S YRYPFV+D + LCY+L P
Sbjct 339 LSPISAQVLRSW-NYKHESDLSNCAEYSRNIVPGSNRNSKYRYPFVYDESEKLCYILYSP 397
Query 148 LQRLMGGK 155
+Q G K
Sbjct 398 MQYNQGVK 405
> bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845
apical merozoite antigen 1
Length=605
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 59/131 (45%), Gaps = 18/131 (13%)
Query 36 RCPVFGKNIEFYQPLDSDLYKNDFLENVPTEEAAAAAKPLP-GGFNNNFLMKDK-KPFSP 93
+CPV GK I+ D FL+ + +E+ + P ++N + K + S
Sbjct 135 KCPVVGKIIDLGNGAD-------FLDPISSEDPSYRGLAFPETAVDSNIPTQPKTRGSSS 187
Query 94 MSVAQLNSYPQLKAR------TGLGKCAE-MSYLTTAAGSS--YRYPFVFDSKKDLCYLL 144
++ A+L+ R + C+E S L A+ + YRYPFVFDS +CY+L
Sbjct 188 VTAAKLSPVSAKDLRRWGYEGNDVANCSEYASNLIPASDKTTKYRYPFVFDSDNQMCYIL 247
Query 145 LVPLQRLMGGK 155
+Q G +
Sbjct 248 YSAIQYNQGNR 258
> hsa:23193 GANAB, G2AN, GLUII, KIAA0088; glucosidase, alpha;
neutral AB (EC:3.2.1.84); K05546 alpha 1,3-glucosidase [EC:3.2.1.84]
Length=944
Score = 33.5 bits (75), Expect = 0.29, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 30/68 (44%), Gaps = 2/68 (2%)
Query 64 PTEEAAAAAKPLPGGFNNNF-LMKDKKPFSPMSVAQLNSYPQLKARTGLGKCAE-MSYLT 121
P E A K PG + F D KP+ PMSV S P ++ G+ + A+ +
Sbjct 211 PEETQGKAEKDEPGAWEETFKTHSDSKPYGPMSVGLDFSLPGMEHVYGIPEHADNLRLKV 270
Query 122 TAAGSSYR 129
T G YR
Sbjct 271 TEGGEPYR 278
> xla:495341 mtmr10-a, mtmr10; myotubularin related protein 10
Length=765
Score = 32.3 bits (72), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 37/75 (49%), Gaps = 6/75 (8%)
Query 48 QPLDSDLYKNDFLENVPT--EEAAAAAKPLPGGFNNNFLMKDKKPFSPMSVAQLNSYPQL 105
PL SD+YK D NVP+ + AA +K N F ++K S + AQ +
Sbjct 309 HPLRSDVYKCDLDHNVPSIQDIQAAFSKLRQLCVNEPFDETEEKWLSSLENAQWLEF--- 365
Query 106 KARTGLGKCAEMSYL 120
RT L + AE++Y+
Sbjct 366 -VRTFLKQAAELAYV 379
> hsa:118987 PDZD8, FLJ25412, FLJ34427, PDZK8, bA129M16.2; PDZ
domain containing 8
Length=1154
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 28/129 (21%), Positives = 54/129 (41%), Gaps = 16/129 (12%)
Query 7 FEKNQTRMEYQETATAAEGPRQLLLLQCRRCPVFGK-----NIEFYQPLDSDLYKNDFLE 61
FE+++ + Q+ A EG ++ LL+C R +FG N+ L S +++
Sbjct 304 FEEDEEHIHIQQWALT-EGRLKVTLLECSRLLIFGSYDREANVHCTLELSSSVWEEKQRS 362
Query 62 NVPTEEAAAAAKPLPG----------GFNNNFLMKDKKPFSPMSVAQLNSYPQLKARTGL 111
++ T E G G+ + +++ P SP ++A L +L A G+
Sbjct 363 SIKTVELIKGNLQSVGLTLRLVQSTDGYAGHVIIETVAPNSPAAIADLQRGDRLIAIGGV 422
Query 112 GKCAEMSYL 120
+ + L
Sbjct 423 KITSTLQVL 431
> mmu:18186 Nrp1, C530029I03, NP-1, NPN-1, Npn1, Nrp; neuropilin
1; K06724 neuropilin 1
Length=923
Score = 30.8 bits (68), Expect = 2.0, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 66 EEAAAAAKPLPGGFNNNFLMKDKKPFSPMSVAQLNSYPQLKARTGLGKCAEM 117
+++ AK G NNN+ + + FSP+S + YP+ +GLG E+
Sbjct 531 DDSKRKAKSFEG--NNNYDTPELRTFSPLSTRFIRIYPERATHSGLGLRMEL 580
> ath:AT4G34280 transducin family protein / WD-40 repeat family
protein
Length=783
Score = 30.8 bits (68), Expect = 2.1, Method: Composition-based stats.
Identities = 15/68 (22%), Positives = 32/68 (47%), Gaps = 5/68 (7%)
Query 49 PLDSDLYKNDFLENVPTEEAAAAAKPLPGGFNNNFLMKD-----KKPFSPMSVAQLNSYP 103
P++ L+ DF +P ++ + + G N LM++ K P+ P V+ ++++
Sbjct 257 PVEETLHSMDFSYKIPEQDDLDSHVSVSAGLNGEVLMREKVRRGKMPYQPKDVSPVDTFT 316
Query 104 QLKARTGL 111
+ GL
Sbjct 317 RQFGNVGL 324
> pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane
antigen 1, AMA1; K13845 apical merozoite antigen 1
Length=622
Score = 30.4 bits (67), Expect = 2.2, Method: Composition-based stats.
Identities = 30/124 (24%), Positives = 49/124 (39%), Gaps = 26/124 (20%)
Query 36 RCPVFGKNIEFYQPLDSDLYKNDFLENVPTEEAAAAAKPLPGGFNNNFLMKDKKPF-SPM 94
+CPVFGK I +EN T A + F +P SPM
Sbjct 148 KCPVFGKGI--------------IIENSNTTFLTPVATGNQYLKDGGFAFPPTEPLMSPM 193
Query 95 SVAQLNSYPQLKARTGLGKCAEMSYLTTAAG---------SSYRYPFVFDSKKDLCYLLL 145
++ ++ + K + E++ + AG S+Y+YP V+D K C++L
Sbjct 194 TLDEMRHF--YKDNKYVKNLDELTLCSRHAGNMIPDNDKNSNYKYPAVYDDKDKKCHILY 251
Query 146 VPLQ 149
+ Q
Sbjct 252 IAAQ 255
> cel:T21C12.3 hypothetical protein
Length=145
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 85 MKDKKPFSPMSVAQLNSYPQLKARTGLGKCAEMSYLTTA 123
++DK+ PMS+ +L P LK R+G K ++ Y + A
Sbjct 54 LRDKRGVDPMSIPRLIKEPPLKKRSGDFKRGDVVYPSAA 92
> mmu:319480 Itga11, 4732459H24Rik; integrin alpha 11; K06587
integrin alpha 11
Length=1188
Score = 30.0 bits (66), Expect = 3.6, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 4/60 (6%)
Query 40 FGKNIEFYQPLDSDLYKNDFLENVPTEEAAAAAKPLPGGFNNNFLMKDKKPFSPMSVAQL 99
FG I Q L+ D Y ND + P E++ A + GF N L KKP ++ ++L
Sbjct 542 FGSCIASVQDLNQDSY-NDVVVGAPLEDSHRGAIYIFHGFQTNIL---KKPMQRITASEL 597
> dre:100333537 ephb1; EPH receptor B1; K05110 Eph receptor B1
[EC:2.7.10.1]
Length=945
Score = 29.3 bits (64), Expect = 5.7, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 6/71 (8%)
Query 8 EKNQTRMEYQETATAAEG--PRQLLLLQCRRCPV--FGK--NIEFYQPLDSDLYKNDFLE 61
E N T+M Q +G P + ++Q R V FGK + +Q L D YK++ E
Sbjct 437 EVNSTQMRSQTNTARVDGLRPGTMYVMQVRARTVAGFGKYSSKMCFQTLTDDDYKSELRE 496
Query 62 NVPTEEAAAAA 72
+P +AAA
Sbjct 497 QLPLIAGSAAA 507
> sce:YOR227W HER1; Her1p
Length=1246
Score = 28.9 bits (63), Expect = 7.2, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 22/47 (46%), Gaps = 0/47 (0%)
Query 63 VPTEEAAAAAKPLPGGFNNNFLMKDKKPFSPMSVAQLNSYPQLKART 109
VPT+ A+ +PL G NN L K S V+Q P + R+
Sbjct 54 VPTKTIASPQRPLSGQNVNNELSNSKPAVSAEKVSQQGQVPTRRTRS 100
Lambda K H
0.319 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3516928200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40