bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7212_orf1
Length=102
Score E
Sequences producing significant alignments: (Bits) Value
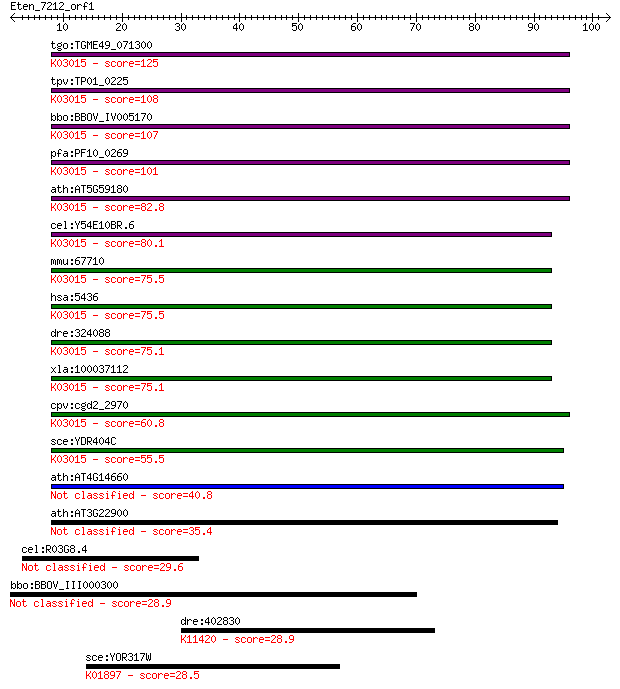
tgo:TGME49_071300 DNA-directed RNA polymerase II subunit RPB7,... 125 2e-29
tpv:TP01_0225 DNA-directed RNA polymerase II subunit Rpb7; K03... 108 3e-24
bbo:BBOV_IV005170 23.m05994; DNA-directed RNA polymerase II (E... 107 8e-24
pfa:PF10_0269 DNA-directed RNA polymerase II, putative; K03015... 101 5e-22
ath:AT5G59180 NRPB7; NRPB7; DNA-directed RNA polymerase/ RNA b... 82.8 3e-16
cel:Y54E10BR.6 rpb-7; RNA Polymerase II (B) subunit family mem... 80.1 2e-15
mmu:67710 Polr2g, 2410046K11Rik, A230108L04Rik, C76415, RBP7, ... 75.5 4e-14
hsa:5436 POLR2G, MGC138367, MGC138369, RPB19, RPB7, hRPB19, hs... 75.5 4e-14
dre:324088 polr2gl, Polr2g, wu:fc18d05, zgc:55297; polymerase ... 75.1 5e-14
xla:100037112 polr2g; polymerase (RNA) II (DNA directed) polyp... 75.1 5e-14
cpv:cgd2_2970 DNA-directed RNA polymerase II ; K03015 DNA-dire... 60.8 1e-09
sce:YDR404C RPB7; B16; K03015 DNA-directed RNA polymerase II s... 55.5 5e-08
ath:AT4G14660 NRPE7; NRPE7; DNA-directed RNA polymerase 40.8 0.001
ath:AT3G22900 NRPD7; NRPD7; DNA-directed RNA polymerase 35.4 0.042
cel:R03G8.4 hypothetical protein 29.6 2.5
bbo:BBOV_III000300 17.m07052; hypothetical protein 28.9 3.7
dre:402830 ehmt1b; euchromatic histone-lysine N-methyltransfer... 28.9 4.3
sce:YOR317W FAA1; Faa1p (EC:6.2.1.3); K01897 long-chain acyl-C... 28.5 5.0
> tgo:TGME49_071300 DNA-directed RNA polymerase II subunit RPB7,
putative (EC:2.7.7.6); K03015 DNA-directed RNA polymerase
II subunit RPB7
Length=207
Score = 125 bits (315), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 56/88 (63%), Positives = 72/88 (81%), Gaps = 0/88 (0%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFSDGSLALRTGGEIR 67
+VLDAVITDVNKLGLFAQCGPLKVFVSRASLPP++ +Q++ A+ +DG + LR +IR
Sbjct 83 EVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPSFVYQDDSAVGCMTDGEVELRVDSDIR 142
Query 68 LKLLGVRFEASQIFAVATTNADYLGPIE 95
L+LLGVR+++ +FA+AT N YLGPI
Sbjct 143 LRLLGVRYDSMNLFAIATINESYLGPIH 170
> tpv:TP01_0225 DNA-directed RNA polymerase II subunit Rpb7; K03015
DNA-directed RNA polymerase II subunit RPB7
Length=175
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 44/88 (50%), Positives = 67/88 (76%), Gaps = 0/88 (0%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFSDGSLALRTGGEIR 67
+VLDA++TDVNKLG FAQ GPLK FVSR S+ P + + + A P +SDG+++++ E+R
Sbjct 83 EVLDAIVTDVNKLGFFAQAGPLKAFVSRNSIHPNFVYDSDSAYPCYSDGTISIKPQSEVR 142
Query 68 LKLLGVRFEASQIFAVATTNADYLGPIE 95
++L G++++ S +FA+AT NAD+LG E
Sbjct 143 MRLQGIKYDNSNLFAIATINADHLGAFE 170
> bbo:BBOV_IV005170 23.m05994; DNA-directed RNA polymerase II
(EC:2.7.7.6); K03015 DNA-directed RNA polymerase II subunit
RPB7
Length=175
Score = 107 bits (268), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 43/88 (48%), Positives = 68/88 (77%), Gaps = 0/88 (0%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFSDGSLALRTGGEIR 67
+VLDA++TDVNKLG FAQ GPLK FVSR ++ P++ + + A P ++DG+++++ E+R
Sbjct 83 EVLDAIVTDVNKLGFFAQAGPLKAFVSRTAIQPSFVYDSDAANPCYTDGTISIKPQTEVR 142
Query 68 LKLLGVRFEASQIFAVATTNADYLGPIE 95
+++ G+R++ S +FA+AT NADYLG E
Sbjct 143 MRIQGIRYDNSNMFAIATVNADYLGAFE 170
> pfa:PF10_0269 DNA-directed RNA polymerase II, putative; K03015
DNA-directed RNA polymerase II subunit RPB7
Length=177
Score = 101 bits (252), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 43/88 (48%), Positives = 63/88 (71%), Gaps = 0/88 (0%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFSDGSLALRTGGEIR 67
+VLDA++TDVNKLG FAQ GPLK+F+SR ++P + + E+ P FS G ++ +R
Sbjct 83 EVLDAIVTDVNKLGFFAQAGPLKIFISRTAIPKYFEYSEDLHYPCFSSGDYNIKPQTTVR 142
Query 68 LKLLGVRFEASQIFAVATTNADYLGPIE 95
+KL G+R++ S +FA+AT N +YLG IE
Sbjct 143 IKLQGIRYDLSNMFAIATINNEYLGCIE 170
> ath:AT5G59180 NRPB7; NRPB7; DNA-directed RNA polymerase/ RNA
binding; K03015 DNA-directed RNA polymerase II subunit RPB7
Length=176
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 40/90 (44%), Positives = 62/90 (68%), Gaps = 3/90 (3%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSF--SDGSLALRTGGE 65
++L+AV+T VNK+G FA+ GP+++FVS+ +P FQ D MP++ SDGS+ ++ E
Sbjct 83 EILEAVVTLVNKMGFFAEAGPVQIFVSKHLIPDDMEFQAGD-MPNYTTSDGSVKIQKECE 141
Query 66 IRLKLLGVRFEASQIFAVATTNADYLGPIE 95
+RLK++G R +A+ IF V T D+LG I
Sbjct 142 VRLKIIGTRVDATAIFCVGTIKDDFLGVIN 171
> cel:Y54E10BR.6 rpb-7; RNA Polymerase II (B) subunit family member
(rpb-7); K03015 DNA-directed RNA polymerase II subunit
RPB7
Length=197
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSF--SDGSLALRTGGE 65
QV+D V+T VNK+G+F GPL F+SR +PP F P + +D + +R E
Sbjct 106 QVVDGVVTQVNKVGIFCDIGPLSCFISRHCIPPDMEFDPNSEKPCYKTNDEANVIRNDDE 165
Query 66 IRLKLLGVRFEASQIFAVATTNADYLG 92
IR+KL+G R +A+ IFA+ T D+LG
Sbjct 166 IRVKLIGTRVDANDIFAIGTLMDDFLG 192
> mmu:67710 Polr2g, 2410046K11Rik, A230108L04Rik, C76415, RBP7,
Rpo2-7l; polymerase (RNA) II (DNA directed) polypeptide G
(EC:2.7.7.6); K03015 DNA-directed RNA polymerase II subunit
RPB7
Length=172
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFS--DGSLALRTGGE 65
+V+DAV+T VNK+GLF + GP+ F+SR S+P F P + D + ++ E
Sbjct 83 EVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFDPNSNPPCYKTMDEDIVIQQDDE 142
Query 66 IRLKLLGVRFEASQIFAVATTNADYLG 92
IRLK++G R + + IFA+ + DYLG
Sbjct 143 IRLKIVGTRVDKNDIFAIGSLMDDYLG 169
> hsa:5436 POLR2G, MGC138367, MGC138369, RPB19, RPB7, hRPB19,
hsRPB7; polymerase (RNA) II (DNA directed) polypeptide G (EC:2.7.7.6);
K03015 DNA-directed RNA polymerase II subunit RPB7
Length=172
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFS--DGSLALRTGGE 65
+V+DAV+T VNK+GLF + GP+ F+SR S+P F P + D + ++ E
Sbjct 83 EVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFDPNSNPPCYKTMDEDIVIQQDDE 142
Query 66 IRLKLLGVRFEASQIFAVATTNADYLG 92
IRLK++G R + + IFA+ + DYLG
Sbjct 143 IRLKIVGTRVDKNDIFAIGSLMDDYLG 169
> dre:324088 polr2gl, Polr2g, wu:fc18d05, zgc:55297; polymerase
(RNA) II (DNA directed) polypeptide G-like (EC:2.7.7.6); K03015
DNA-directed RNA polymerase II subunit RPB7
Length=172
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFS--DGSLALRTGGE 65
+V+DAV+T VNK+GLF + GP+ F+SR S+P F P + D + ++ E
Sbjct 83 EVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFDPNSNPPCYKTVDEDVVIQQDDE 142
Query 66 IRLKLLGVRFEASQIFAVATTNADYLG 92
IRLK++G R + + IFA+ + DYLG
Sbjct 143 IRLKIVGTRVDKNDIFAIGSLMDDYLG 169
> xla:100037112 polr2g; polymerase (RNA) II (DNA directed) polypeptide
G; K03015 DNA-directed RNA polymerase II subunit RPB7
Length=172
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 53/87 (60%), Gaps = 2/87 (2%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFS--DGSLALRTGGE 65
+V+DAV+T VNK+GLF + GP+ F+SR S+P F P + D + ++ E
Sbjct 83 EVVDAVVTQVNKVGLFTEIGPMSCFISRHSIPSEMEFDPNSNPPCYKTVDEDVVIQQDDE 142
Query 66 IRLKLLGVRFEASQIFAVATTNADYLG 92
IRLK++G R + + IFA+ + DYLG
Sbjct 143 IRLKIVGTRVDKNDIFAIGSLMDDYLG 169
> cpv:cgd2_2970 DNA-directed RNA polymerase II ; K03015 DNA-directed
RNA polymerase II subunit RPB7
Length=178
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/94 (35%), Positives = 56/94 (59%), Gaps = 8/94 (8%)
Query 8 QVLDAVITDVNKLGLFAQCGPLK-VFVSRASLPPTYTFQ-----EEDAMPSFSDGSLALR 61
+V+D V+ VN+LG+ CGPLK VFVS+++LP ++ + + + + ++
Sbjct 83 EVVDGVVESVNELGVIVDCGPLKRVFVSQSALPENMVYKSGIDGQSSRIYIDTKTQIQIK 142
Query 62 TGGEIRLKLLGVRFEASQIFAVATTNADYLGPIE 95
++RL+L GV E + AVA N+D+LGPI+
Sbjct 143 QNSQVRLRLRGVNNET--LSAVAEMNSDFLGPID 174
> sce:YDR404C RPB7; B16; K03015 DNA-directed RNA polymerase II
subunit RPB7
Length=171
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 43/87 (49%), Gaps = 0/87 (0%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFSDGSLALRTGGEIR 67
+V+D + ++ G Q GP+KVFV++ +P TF PS+ + IR
Sbjct 85 EVVDGTVVSCSQHGFEVQVGPMKVFVTKHLMPQDLTFNAGSNPPSYQSSEDVITIKSRIR 144
Query 68 LKLLGVRFEASQIFAVATTNADYLGPI 94
+K+ G + S I A+ + DYLG I
Sbjct 145 VKIEGCISQVSSIHAIGSIKEDYLGAI 171
> ath:AT4G14660 NRPE7; NRPE7; DNA-directed RNA polymerase
Length=178
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 53/91 (58%), Gaps = 6/91 (6%)
Query 8 QVLDAVITDVNKLGLFAQCGPLK-VFVSRASLPPTYTFQEEDAMPSF-SDGSLALRTGGE 65
+++ V+ V K G+F +CGP++ V++S +P E+ P F ++ + ++
Sbjct 87 EIIHGVVHKVLKHGVFMRCGPIENVYLSYTKMPDYKYIPGEN--PIFMNEKTSRIQVETT 144
Query 66 IRLKLLGVRF-EASQIF-AVATTNADYLGPI 94
+R+ ++G+++ E + F A+A+ DYLGP+
Sbjct 145 VRVVVIGIKWMEVEREFQALASLEGDYLGPL 175
> ath:AT3G22900 NRPD7; NRPD7; DNA-directed RNA polymerase
Length=174
Score = 35.4 bits (80), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 46/89 (51%), Gaps = 4/89 (4%)
Query 8 QVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSFSDGSLA-LRTGGEI 66
+++ V+ V+K G+F + GP ++ P Y F + P F + ++ ++ G +
Sbjct 86 EIVHGVVHKVHKTGVFLKSGPYEIIYLSHMKMPGYEFIPGEN-PFFMNQYMSRIQIGARV 144
Query 67 RLKLLGVRF-EASQIF-AVATTNADYLGP 93
R +L + EA + F A+A+ + D LGP
Sbjct 145 RFVVLDTEWREAEKDFMALASIDGDNLGP 173
> cel:R03G8.4 hypothetical protein
Length=786
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 23/31 (74%), Gaps = 1/31 (3%)
Query 3 LSQWSQVLDAV-ITDVNKLGLFAQCGPLKVF 32
+++ S++L+ + I +VN + +F++CGP K F
Sbjct 647 INEASRILEGIQIEEVNDIDIFSKCGPFKEF 677
> bbo:BBOV_III000300 17.m07052; hypothetical protein
Length=420
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 27/75 (36%), Positives = 33/75 (44%), Gaps = 12/75 (16%)
Query 1 STLSQWSQVLDAVITDVNKLGLFAQCGPLKVFVSRASLPPT-----YTFQEEDAMPSFSD 55
STL + + LD +NKLGLF K VS PT FQ E+ PS+S
Sbjct 58 STLEELFRRLDDTHGHLNKLGLF------KSVVSNVDRGPTPGDIDVVFQFEEKTPSYSL 111
Query 56 G-SLALRTGGEIRLK 69
G + R I LK
Sbjct 112 GVTTNQRAEANIELK 126
> dre:402830 ehmt1b; euchromatic histone-lysine N-methyltransferase
1b; K11420 euchromatic histone-lysine N-methyltransferase
[EC:2.1.1.43]
Length=1278
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 30 KVFVSRASLPPTYTFQEEDAMPSFSDGSLALRTGGEIRLKLLG 72
+V +++A T +PS S+G TGG RL +LG
Sbjct 610 EVTIAKADTTSTVPITHSSCIPSTSEGKADTTTGGSTRLSVLG 652
> sce:YOR317W FAA1; Faa1p (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=700
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 26/44 (59%), Gaps = 1/44 (2%)
Query 14 ITDVNKLGLFAQCGPLKVFVSRASLPPTYTFQEEDAMPSF-SDG 56
+ DV +LG FA+ +V+++ A++ P Y EE+ + SDG
Sbjct 489 LVDVEELGYFAKNNQGEVWITGANVTPEYYKNEEETSQALTSDG 532
Lambda K H
0.318 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2031832220
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40