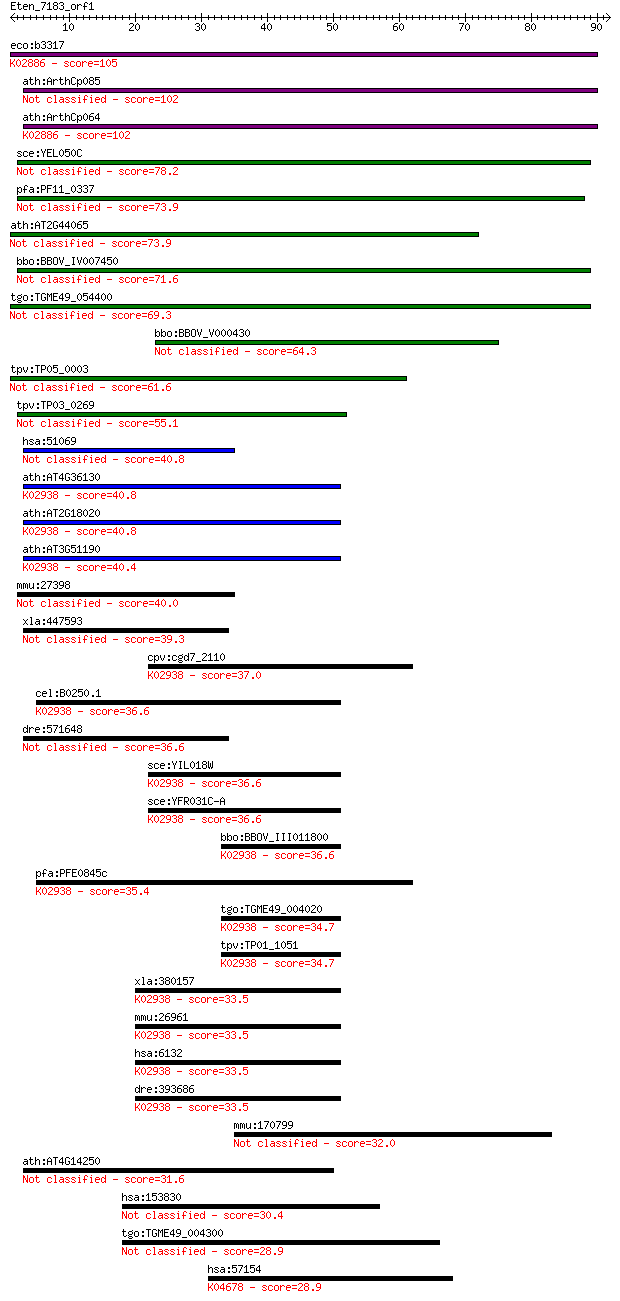
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7183_orf1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
eco:b3317 rplB, ECK3304, JW3279; 50S ribosomal subunit protein... 105 3e-23
ath:ArthCp085 rpl2; 50S ribosomal protein L2 102 2e-22
ath:ArthCp064 rpl2; 50S ribosomal protein L2; K02886 large sub... 102 2e-22
sce:YEL050C RML2; Mitochondrial ribosomal protein of the large... 78.2 5e-15
pfa:PF11_0337 mitochondrial ribosomal protein L2 precursor 73.9 1e-13
ath:AT2G44065 ribosomal protein L2 family protein 73.9 1e-13
bbo:BBOV_IV007450 23.m05961; ribosomal protein L2 71.6 5e-13
tgo:TGME49_054400 50S ribosomal protein l2, putative 69.3 3e-12
bbo:BBOV_V000430 ribosomal protein L2 64.3 9e-11
tpv:TP05_0003 rpl2; ribosomal protein L2 61.6 5e-10
tpv:TP03_0269 60S ribosomal protein L2 55.1 5e-08
hsa:51069 MRPL2, MRP-L14, RPML14; mitochondrial ribosomal prot... 40.8 0.001
ath:AT4G36130 60S ribosomal protein L8 (RPL8C); K02938 large s... 40.8 0.001
ath:AT2G18020 EMB2296 (embryo defective 2296); structural cons... 40.8 0.001
ath:AT3G51190 structural constituent of ribosome; K02938 large... 40.4 0.001
mmu:27398 Mrpl2, CGI-22, MRP-L14, Rpml14; mitochondrial riboso... 40.0 0.002
xla:447593 MGC84466 protein 39.3 0.003
cpv:cgd7_2110 60S ribosomal proteins L8/L2 ; K02938 large subu... 37.0 0.013
cel:B0250.1 rpl-2; Ribosomal Protein, Large subunit family mem... 36.6 0.018
dre:571648 im:6908224 36.6 0.021
sce:YIL018W RPL2B, LOT2, RPL5A; Rpl2bp; K02938 large subunit r... 36.6 0.021
sce:YFR031C-A RPL2A, RPL5B; Rpl2ap; K02938 large subunit ribos... 36.6 0.021
bbo:BBOV_III011800 17.m08006; ribosomal protein L2 family prot... 36.6 0.021
pfa:PFE0845c 60S ribosomal protein L8, putative; K02938 large ... 35.4 0.049
tgo:TGME49_004020 60S ribosomal protein L8, putative ; K02938 ... 34.7 0.072
tpv:TP01_1051 60S ribosomal protein L8; K02938 large subunit r... 34.7 0.075
xla:380157 rpl8, MGC53203; ribosomal protein L8; K02938 large ... 33.5 0.16
mmu:26961 Rpl8; ribosomal protein L8; K02938 large subunit rib... 33.5 0.18
hsa:6132 RPL8; ribosomal protein L8; K02938 large subunit ribo... 33.5 0.18
dre:393686 rpl8, MGC73105, zgc:73105, zgc:77641; ribosomal pro... 33.5 0.18
mmu:170799 Rtkn2, B130039D23Rik, MGC189880, Mbf, Plekhk1; rhot... 32.0 0.48
ath:AT4G14250 structural constituent of ribosome 31.6 0.67
hsa:153830 RNF145, DKFZp686M11215, FLJ31951, FLJ44310; ring fi... 30.4 1.5
tgo:TGME49_004300 hypothetical protein 28.9 3.6
hsa:57154 SMURF1, KIAA1625; SMAD specific E3 ubiquitin protein... 28.9 4.3
> eco:b3317 rplB, ECK3304, JW3279; 50S ribosomal subunit protein
L2; K02886 large subunit ribosomal protein L2
Length=273
Score = 105 bits (262), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 56/89 (62%), Positives = 64/89 (71%), Gaps = 2/89 (2%)
Query 1 GECMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKA 60
+C AT+GEVGN+EH L +GKAG R G RP VRG+ MNPVDHPHGGGEG+N K
Sbjct 186 ADCRATLGEVGNAEHMLRVLGKAGAARWRGVRPTVRGTAMNPVDHPHGGGEGRN--FGKH 243
Query 61 PLTPWGKPALGVKTRGKKTTDKFIVRRRN 89
P+TPWG G KTR K TDKFIVRRR+
Sbjct 244 PVTPWGVQTKGKKTRSNKRTDKFIVRRRS 272
> ath:ArthCp085 rpl2; 50S ribosomal protein L2
Length=274
Score = 102 bits (255), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 51/88 (57%), Positives = 66/88 (75%), Gaps = 1/88 (1%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAPL 62
C AT+G+VGN + +G+AG +G+RP VRG VMNPVDHPHGGGEG+ P+GRK P+
Sbjct 186 CSATVGQVGNVGVNQKSLGRAGSKCWLGKRPVVRGVVMNPVDHPHGGGEGRAPIGRKKPV 245
Query 63 TPWGKPALGVKTRG-KKTTDKFIVRRRN 89
TPWG PALG +TR KK ++ I+RRR+
Sbjct 246 TPWGYPALGRRTRKRKKYSETLILRRRS 273
> ath:ArthCp064 rpl2; 50S ribosomal protein L2; K02886 large subunit
ribosomal protein L2
Length=274
Score = 102 bits (255), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 51/88 (57%), Positives = 66/88 (75%), Gaps = 1/88 (1%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAPL 62
C AT+G+VGN + +G+AG +G+RP VRG VMNPVDHPHGGGEG+ P+GRK P+
Sbjct 186 CSATVGQVGNVGVNQKSLGRAGSKCWLGKRPVVRGVVMNPVDHPHGGGEGRAPIGRKKPV 245
Query 63 TPWGKPALGVKTRG-KKTTDKFIVRRRN 89
TPWG PALG +TR KK ++ I+RRR+
Sbjct 246 TPWGYPALGRRTRKRKKYSETLILRRRS 273
> sce:YEL050C RML2; Mitochondrial ribosomal protein of the large
subunit, has similarity to E. coli L2 ribosomal protein;
fat21 mutant allele causes inability to utilize oleate and may
interfere with activity of the Adr1p transcription factor
Length=393
Score = 78.2 bits (191), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 43/87 (49%), Positives = 55/87 (63%), Gaps = 1/87 (1%)
Query 2 ECMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAP 61
E +ATIG V N +H +GKAGR+R +G RP VRG MN DHPHGGG GK+ K
Sbjct 300 EAVATIGVVSNIDHQNRSLGKAGRSRWLGIRPTVRGVAMNKCDHPHGGGRGKSK-SNKLS 358
Query 62 LTPWGKPALGVKTRGKKTTDKFIVRRR 88
++PWG+ A G KTR K ++ V+ R
Sbjct 359 MSPWGQLAKGYKTRRGKNQNRMKVKDR 385
> pfa:PF11_0337 mitochondrial ribosomal protein L2 precursor
Length=333
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 42/86 (48%), Positives = 50/86 (58%), Gaps = 1/86 (1%)
Query 2 ECMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAP 61
+C ATIG+V N E + +GKAG NR +G+RP VRG MNP HPHGGG K R
Sbjct 235 QCWATIGQVSNLERHMRILGKAGVNRWLGKRPVVRGVAMNPSKHPHGGGTSKKHTKR-PK 293
Query 62 LTPWGKPALGVKTRGKKTTDKFIVRR 87
+ WG G KTR KK I+RR
Sbjct 294 CSLWGICRDGYKTRSKKKPLGLIIRR 319
> ath:AT2G44065 ribosomal protein L2 family protein
Length=214
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 39/75 (52%), Positives = 49/75 (65%), Gaps = 5/75 (6%)
Query 1 GECMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPV---- 56
+C ATIG V N H ++ KAG++R +G RP VRG MNP DHPHGGGEGK+
Sbjct 120 AKCRATIGTVSNPSHGTKKLYKAGQSRWLGIRPKVRGVAMNPCDHPHGGGEGKSKSSGSR 179
Query 57 GRKAPLTPWGKPALG 71
GR + ++PWGKP G
Sbjct 180 GRTS-VSPWGKPCKG 193
> bbo:BBOV_IV007450 23.m05961; ribosomal protein L2
Length=332
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 42/88 (47%), Positives = 55/88 (62%), Gaps = 3/88 (3%)
Query 2 ECMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAP 61
+C AT+G+V N EH+ + KAG R++G RP VRG MNP HPHGGG K+ G K P
Sbjct 232 DCWATLGQVSNIEHNKRILKKAGTRRNLGWRPSVRGIAMNPNAHPHGGGNNKS--GTKRP 289
Query 62 -LTPWGKPALGVKTRGKKTTDKFIVRRR 88
+ WG G+KTR K+ FIV+R+
Sbjct 290 KCSVWGVCRSGLKTRSKQKHLGFIVKRK 317
> tgo:TGME49_054400 50S ribosomal protein l2, putative
Length=380
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 44/88 (50%), Positives = 54/88 (61%), Gaps = 1/88 (1%)
Query 1 GECMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKA 60
+C AT+G+V N+ H+ GKAG + MG RP RG MNPVDHPHGGG GK + R
Sbjct 280 ADCWATVGQVSNAAHAERIRGKAGVSYWMGERPRTRGKAMNPVDHPHGGGTGKKGLKR-P 338
Query 61 PLTPWGKPALGVKTRGKKTTDKFIVRRR 88
P++ WG G KTR KK IVRR+
Sbjct 339 PVSKWGILCKGYKTRAKKKPLGLIVRRK 366
> bbo:BBOV_V000430 ribosomal protein L2
Length=242
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 30/52 (57%), Positives = 35/52 (67%), Gaps = 0/52 (0%)
Query 23 AGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAPLTPWGKPALGVKT 74
AG +GRRP VRG+ MN DHPHGGGEGK P+GRK + G+ GVKT
Sbjct 186 AGYKIKLGRRPKVRGTAMNACDHPHGGGEGKAPIGRKTIYSFTGRKCKGVKT 237
> tpv:TP05_0003 rpl2; ribosomal protein L2
Length=246
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 32/61 (52%), Positives = 37/61 (60%), Gaps = 4/61 (6%)
Query 1 GECMATIGEVGNSEHSLVQI-GKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRK 59
+C A GNSE + KAG R G RP VRGS MN DHPHGGGEGK P+GR+
Sbjct 166 NKCYAV---YGNSEKVISNFKQKAGTERLFGNRPKVRGSAMNACDHPHGGGEGKAPIGRR 222
Query 60 A 60
+
Sbjct 223 S 223
> tpv:TP03_0269 60S ribosomal protein L2
Length=275
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 26/50 (52%), Positives = 34/50 (68%), Gaps = 0/50 (0%)
Query 2 ECMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGE 51
+C ATIG++ N +H+ + KAG R+MG RP VRG MNP HPHGG +
Sbjct 226 DCWATIGQISNIKHNERILKKAGTRRNMGWRPVVRGIAMNPNSHPHGGNK 275
> hsa:51069 MRPL2, MRP-L14, RPML14; mitochondrial ribosomal protein
L2
Length=305
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/32 (56%), Positives = 25/32 (78%), Gaps = 0/32 (0%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRPH 34
C+AT+G V N +H+ IGKAGRNR +G+RP+
Sbjct 240 CVATVGRVSNVDHNKRVIGKAGRNRWLGKRPN 271
> ath:AT4G36130 60S ribosomal protein L8 (RPL8C); K02938 large
subunit ribosomal protein L8e
Length=258
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/53 (45%), Positives = 31/53 (58%), Gaps = 5/53 (9%)
Query 3 CMATIGEV---GNSEHSLVQIGKAGRNRHMGRR--PHVRGSVMNPVDHPHGGG 50
C A IG+V G +E +++ G A + R P VRG MNPV+HPHGGG
Sbjct 162 CRAMIGQVAGGGRTEKPMLKAGNAYHKYRVKRNCWPKVRGVAMNPVEHPHGGG 214
> ath:AT2G18020 EMB2296 (embryo defective 2296); structural constituent
of ribosome; K02938 large subunit ribosomal protein
L8e
Length=258
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/53 (45%), Positives = 31/53 (58%), Gaps = 5/53 (9%)
Query 3 CMATIGEV---GNSEHSLVQIGKAGRNRHMGRR--PHVRGSVMNPVDHPHGGG 50
C A IG+V G +E +++ G A + R P VRG MNPV+HPHGGG
Sbjct 162 CRAMIGQVAGGGRTEKPMLKAGNAYHKYRVKRNSWPKVRGVAMNPVEHPHGGG 214
> ath:AT3G51190 structural constituent of ribosome; K02938 large
subunit ribosomal protein L8e
Length=260
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 34/54 (62%), Gaps = 7/54 (12%)
Query 3 CMATIGEV---GNSEHSLVQIGKAGRNRHMGRR---PHVRGSVMNPVDHPHGGG 50
C A IG+V G +E L++ G A +++ +R P VRG MNPV+HPHGGG
Sbjct 163 CRAMIGQVAGGGRTEKPLLKAGNA-YHKYKAKRNCWPVVRGVAMNPVEHPHGGG 215
> mmu:27398 Mrpl2, CGI-22, MRP-L14, Rpml14; mitochondrial ribosomal
protein L2
Length=306
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/33 (54%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 2 ECMATIGEVGNSEHSLVQIGKAGRNRHMGRRPH 34
C AT+G V N H+ IGKAGRNR +G+RP+
Sbjct 240 SCTATVGRVSNVNHNQRVIGKAGRNRWLGKRPN 272
> xla:447593 MGC84466 protein
Length=294
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/31 (58%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRP 33
C+AT+G V N +H+ IGKAGRNR +G RP
Sbjct 228 CVATVGRVSNIDHNKRIIGKAGRNRWLGIRP 258
> cpv:cgd7_2110 60S ribosomal proteins L8/L2 ; K02938 large subunit
ribosomal protein L8e
Length=261
Score = 37.0 bits (84), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 24/50 (48%), Positives = 27/50 (54%), Gaps = 10/50 (20%)
Query 22 KAGRNRHMGRR-----PHVRGSVMNPVDHPHGGGEGK-----NPVGRKAP 61
KAG N H + P VRG MNPVDHPHGGG + + V R AP
Sbjct 181 KAGNNYHKYKVKRNCWPKVRGVSMNPVDHPHGGGNHQHIGHPSTVSRSAP 230
> cel:B0250.1 rpl-2; Ribosomal Protein, Large subunit family member
(rpl-2); K02938 large subunit ribosomal protein L8e
Length=260
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 28/51 (54%), Gaps = 5/51 (9%)
Query 5 ATIGEVGNSEHSLVQIGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
A IG V + + KAGR+ H + P VRG MNPV+HPHGGG
Sbjct 164 AMIGLVAGGGRTDKPLLKAGRSYHKYKAKRNSWPRVRGVAMNPVEHPHGGG 214
> dre:571648 im:6908224
Length=294
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 17/31 (54%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRP 33
C+AT+G V N +H+ IGKAG NR +G RP
Sbjct 229 CVATVGRVSNVDHNKRIIGKAGTNRWLGIRP 259
> sce:YIL018W RPL2B, LOT2, RPL5A; Rpl2bp; K02938 large subunit
ribosomal protein L8e
Length=254
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 20/34 (58%), Positives = 20/34 (58%), Gaps = 5/34 (14%)
Query 22 KAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
KAGR H R P RG MNPVDHPHGGG
Sbjct 181 KAGRAFHKYRLKRNSWPKTRGVAMNPVDHPHGGG 214
> sce:YFR031C-A RPL2A, RPL5B; Rpl2ap; K02938 large subunit ribosomal
protein L8e
Length=254
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 20/34 (58%), Positives = 20/34 (58%), Gaps = 5/34 (14%)
Query 22 KAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
KAGR H R P RG MNPVDHPHGGG
Sbjct 181 KAGRAFHKYRLKRNSWPKTRGVAMNPVDHPHGGG 214
> bbo:BBOV_III011800 17.m08006; ribosomal protein L2 family protein;
K02938 large subunit ribosomal protein L8e
Length=257
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 15/18 (83%), Positives = 15/18 (83%), Gaps = 0/18 (0%)
Query 33 PHVRGSVMNPVDHPHGGG 50
P VRG MNPVDHPHGGG
Sbjct 197 PRVRGVTMNPVDHPHGGG 214
> pfa:PFE0845c 60S ribosomal protein L8, putative; K02938 large
subunit ribosomal protein L8e
Length=260
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 32/67 (47%), Gaps = 10/67 (14%)
Query 5 ATIGEVGNSEHSLVQIGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGGEGK-----N 54
A +G VG I KAG H R P VRG MNPV+HPHGGG + +
Sbjct 164 AMVGVVGAGGRIDKPILKAGVAHHKYRVKRNCWPKVRGVAMNPVEHPHGGGNHQHIGHPS 223
Query 55 PVGRKAP 61
V R AP
Sbjct 224 TVSRSAP 230
> tgo:TGME49_004020 60S ribosomal protein L8, putative ; K02938
large subunit ribosomal protein L8e
Length=260
Score = 34.7 bits (78), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 14/18 (77%), Positives = 15/18 (83%), Gaps = 0/18 (0%)
Query 33 PHVRGSVMNPVDHPHGGG 50
P VRG MNPV+HPHGGG
Sbjct 197 PKVRGVAMNPVEHPHGGG 214
> tpv:TP01_1051 60S ribosomal protein L8; K02938 large subunit
ribosomal protein L8e
Length=258
Score = 34.7 bits (78), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 14/18 (77%), Positives = 15/18 (83%), Gaps = 0/18 (0%)
Query 33 PHVRGSVMNPVDHPHGGG 50
P VRG MNPV+HPHGGG
Sbjct 197 PKVRGVAMNPVEHPHGGG 214
> xla:380157 rpl8, MGC53203; ribosomal protein L8; K02938 large
subunit ribosomal protein L8e
Length=257
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 21/36 (58%), Gaps = 5/36 (13%)
Query 20 IGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
I KAGR H + P VRG MNPV+HP GGG
Sbjct 179 ILKAGRAYHKYKAKRNCWPRVRGVAMNPVEHPFGGG 214
> mmu:26961 Rpl8; ribosomal protein L8; K02938 large subunit ribosomal
protein L8e
Length=257
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 21/36 (58%), Gaps = 5/36 (13%)
Query 20 IGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
I KAGR H + P VRG MNPV+HP GGG
Sbjct 179 ILKAGRAYHKYKAKRNCWPRVRGVAMNPVEHPFGGG 214
> hsa:6132 RPL8; ribosomal protein L8; K02938 large subunit ribosomal
protein L8e
Length=257
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 21/36 (58%), Gaps = 5/36 (13%)
Query 20 IGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
I KAGR H + P VRG MNPV+HP GGG
Sbjct 179 ILKAGRAYHKYKAKRNCWPRVRGVAMNPVEHPFGGG 214
> dre:393686 rpl8, MGC73105, zgc:73105, zgc:77641; ribosomal protein
L8; K02938 large subunit ribosomal protein L8e
Length=257
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 21/36 (58%), Gaps = 5/36 (13%)
Query 20 IGKAGRNRHMGRR-----PHVRGSVMNPVDHPHGGG 50
I KAGR H + P VRG MNPV+HP GGG
Sbjct 179 ILKAGRAYHKYKAKRNCWPRVRGVAMNPVEHPFGGG 214
> mmu:170799 Rtkn2, B130039D23Rik, MGC189880, Mbf, Plekhk1; rhotekin
2
Length=602
Score = 32.0 bits (71), Expect = 0.48, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 3/48 (6%)
Query 35 VRGSVMNPVDHPHGGGEGKNPVGRKAPLTPWGKPALGVKTRGKKTTDK 82
+ V++P+ P G+ K R+APL P +P +KT+G+ K
Sbjct 474 IEKKVLSPIGEPAPDGKRKK---RRAPLPPTDQPPFCIKTQGRANQSK 518
> ath:AT4G14250 structural constituent of ribosome
Length=724
Score = 31.6 bits (70), Expect = 0.67, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 28/48 (58%), Gaps = 7/48 (14%)
Query 3 CMATIGEVGNSEHSLVQIGKAGRNRHMGRRPHVRG-SVMNPVDHPHGG 49
C IG++ +S + K R+M + VRG ++MNPV+HPHGG
Sbjct 396 CRVMIGQIASSGLT----KKLMIKRNMWAK--VRGVAMMNPVEHPHGG 437
> hsa:153830 RNF145, DKFZp686M11215, FLJ31951, FLJ44310; ring
finger protein 145
Length=693
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 11/39 (28%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 18 VQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPV 56
+Q G N ++ RRP + +P ++PH + +PV
Sbjct 652 IQEGSRDNNEYIARRPDNQEGAFDPKEYPHSAKDEAHPV 690
> tgo:TGME49_004300 hypothetical protein
Length=744
Score = 28.9 bits (63), Expect = 3.6, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query 18 VQIGKAGRNRHMGRRPHVRGSVMNPVDHPHGGGEGKNPVGRKAPLTPW 65
V G+A + R+P V + V+ PH G G+ P R++ T W
Sbjct 404 VAAGRAAQRPQRSRKPTVSLRSVVTVNTPHAGVAGR-PFARRSSFTEW 450
> hsa:57154 SMURF1, KIAA1625; SMAD specific E3 ubiquitin protein
ligase 1; K04678 E3 ubiquitin ligase SMURF1/2 [EC:6.3.2.19]
Length=728
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 31 RRPHVRGSVMNPVDHPHGGGEGKNPVGRKAPLTPWGK 67
R P VRGS+ P + PHG + P G + T G+
Sbjct 213 RNPDVRGSLQTPQNRPHGHQSPELPEGYEQRTTVQGQ 249
Lambda K H
0.315 0.137 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007827920
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40