bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
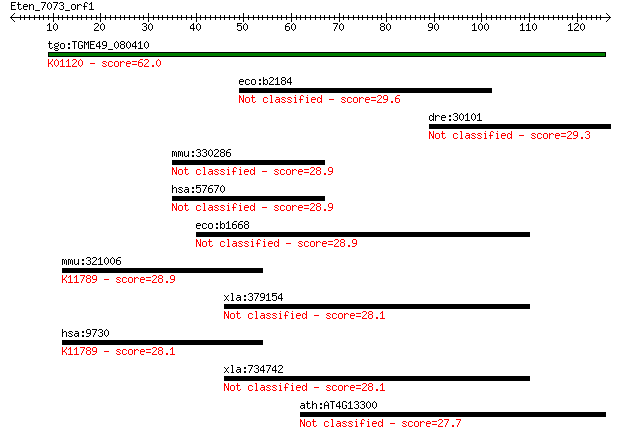
Query= Eten_7073_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC... 62.0 4e-10
eco:b2184 yejH, ECK2178, JW2172, yejI, yejJ; predicted ATP-dep... 29.6 2.4
dre:30101 skia, MGC136741, id:ibd1250; nuclear oncoprotein skia 29.3 3.0
mmu:330286 D630045J12Rik, Kiaa1549, mKIAA1549; RIKEN cDNA D630... 28.9 3.8
hsa:57670 FLJ11731, FLJ38703, FLJ40644; KIAA1549 28.9 3.8
eco:b1668 ydhS, ECK1664, JW1658; conserved protein with FAD/NA... 28.9 4.4
mmu:321006 Vprbp, AI447437, B930007L02Rik, mKIAA0800; Vpr (HIV... 28.9 4.4
xla:379154 MGC52862; similar to tektin 2 (testicular) 28.1 6.3
hsa:9730 VPRBP, DCAF1, KIAA0800, MGC102804; Vpr (HIV-1) bindin... 28.1 6.7
xla:734742 tekt2, MGC130819; tektin 2 (testicular) 28.1 7.0
ath:AT4G13300 TPS13; TPS13 (TERPENOID SYNTHASE 13); cyclase 27.7 8.6
> tgo:TGME49_080410 3',5'-cyclic phosphodiesterase, putative (EC:3.1.4.17
1.6.5.3); K01120 3',5'-cyclic-nucleotide phosphodiesterase
[EC:3.1.4.17]
Length=1085
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 40/118 (33%), Positives = 68/118 (57%), Gaps = 8/118 (6%)
Query 9 LIYFLLAVLVMVLDQAAMIHEHKRRVLFWKLKTADDRLDLLEDQIFHKQKRKRVPVPSTP 68
++ FLL VL +V +A E K+RVLFW L+ +D RL LLE Q+ ++ ++RV TP
Sbjct 594 ILPFLLIVLTLV---SAYYEERKKRVLFWNLRISDSRLKLLEKQVQLRRLKRRVDT-GTP 649
Query 69 LESVIRNL-DLVQGVVLSAQMGEDERARVQQLLTEARHNLTFSDNIYQFNADTCKDGF 125
L+ ++ NL + ++ + + +E + L+ E + LT S+N+YQ A+ + F
Sbjct 650 LDCILGNLSEALEALKRPTPLNREE---IFALMQETINMLTTSENVYQVQAEYVDNDF 704
> eco:b2184 yejH, ECK2178, JW2172, yejI, yejJ; predicted ATP-dependent
DNA or RNA helicase
Length=586
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/62 (37%), Positives = 32/62 (51%), Gaps = 9/62 (14%)
Query 49 LEDQIFH---KQKRKRVPVPSTPLESVIRNLDLVQG------VVLSAQMGEDERARVQQL 99
LE IF K+K V ++SV RNLD QG V ++G+DE ++ QQ+
Sbjct 74 LEADIFAAGLKRKESHGKVVFGSVQSVARNLDAFQGEFSLLIVDECHRIGDDEESQYQQI 133
Query 100 LT 101
LT
Sbjct 134 LT 135
> dre:30101 skia, MGC136741, id:ibd1250; nuclear oncoprotein skia
Length=727
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query 89 GEDERARVQQLLTEARHNLTFSDNIYQFNADTCKDGFP 126
G++E+AR++QLL + + FS N Y+ A D P
Sbjct 274 GKEEKARLEQLLDDIKEKFDFS-NKYKRKASQASDPIP 310
> mmu:330286 D630045J12Rik, Kiaa1549, mKIAA1549; RIKEN cDNA D630045J12
gene
Length=1940
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 21/32 (65%), Gaps = 1/32 (3%)
Query 35 LFWKLKTADDRLDLLEDQIFHKQKRKRVPVPS 66
L+WKL D+LD D + + Q+R+++ +PS
Sbjct 1308 LYWKL-CRTDKLDFQPDTVANIQQRQKLQIPS 1338
> hsa:57670 FLJ11731, FLJ38703, FLJ40644; KIAA1549
Length=1950
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 21/32 (65%), Gaps = 1/32 (3%)
Query 35 LFWKLKTADDRLDLLEDQIFHKQKRKRVPVPS 66
L+WKL D+LD D + + Q+R+++ +PS
Sbjct 1319 LYWKL-CRTDKLDFQPDTVANIQQRQKLQIPS 1349
> eco:b1668 ydhS, ECK1664, JW1658; conserved protein with FAD/NAD(P)-binding
domain
Length=534
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 33/70 (47%), Gaps = 0/70 (0%)
Query 40 KTADDRLDLLEDQIFHKQKRKRVPVPSTPLESVIRNLDLVQGVVLSAQMGEDERARVQQL 99
KT D+ D+ ED + + R V L ++ + VQG+ A++GE V +
Sbjct 464 KTGDEIPDVGEDYTLQQPEDIRGRVAFGALPWLMHDQPFVQGLTACAEIGEAMARAVVKP 523
Query 100 LTEARHNLTF 109
+ AR L+F
Sbjct 524 ASRARRRLSF 533
> mmu:321006 Vprbp, AI447437, B930007L02Rik, mKIAA0800; Vpr (HIV-1)
binding protein; K11789 HIV-1 Vpr-binding protein
Length=1419
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 12 FLLAVLVMVLDQAAMIHEHKRRVLFWKLKTADDRLDLLEDQI 53
F L V LDQ ++ H V++ + ADD DLLE+++
Sbjct 1284 FHLLHTVPALDQCRVVFNHTGTVMYGAMLQADDEDDLLEERM 1325
> xla:379154 MGC52862; similar to tektin 2 (testicular)
Length=430
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 35/65 (53%), Gaps = 1/65 (1%)
Query 46 LDLLEDQIFHKQKRKRVPVPSTPLESVI-RNLDLVQGVVLSAQMGEDERARVQQLLTEAR 104
LD+ + + H++ R+ + + P+E + + +++++GV + Q E LL EAR
Sbjct 110 LDVAIECLTHRESRREIDLVKDPVEDELHKEVEVIEGVRRALQQKISEAFEQLCLLQEAR 169
Query 105 HNLTF 109
H L F
Sbjct 170 HQLNF 174
> hsa:9730 VPRBP, DCAF1, KIAA0800, MGC102804; Vpr (HIV-1) binding
protein; K11789 HIV-1 Vpr-binding protein
Length=1506
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 12 FLLAVLVMVLDQAAMIHEHKRRVLFWKLKTADDRLDLLEDQI 53
F L V LDQ ++ H V++ + ADD DL+E+++
Sbjct 1284 FHLLHTVPALDQCRVVFNHTGTVMYGAMLQADDEDDLMEERM 1325
> xla:734742 tekt2, MGC130819; tektin 2 (testicular)
Length=430
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 35/65 (53%), Gaps = 1/65 (1%)
Query 46 LDLLEDQIFHKQKRKRVPVPSTPLESVI-RNLDLVQGVVLSAQMGEDERARVQQLLTEAR 104
LD+ + + H++ R+ + + P+E + + +++++GV + Q E LL EAR
Sbjct 110 LDVAIECLTHRESRREIDLVKDPVEDELHKEVEVIEGVRRALQQKISEAFEQLCLLQEAR 169
Query 105 HNLTF 109
H L F
Sbjct 170 HQLNF 174
> ath:AT4G13300 TPS13; TPS13 (TERPENOID SYNTHASE 13); cyclase
Length=554
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 36/66 (54%), Gaps = 2/66 (3%)
Query 62 VPVPSTPLESVIRNLDLVQGVV--LSAQMGEDERARVQQLLTEARHNLTFSDNIYQFNAD 119
VP+ + L+++ R +D+++ V L + G+DE ++ + LL ++ +L + + D
Sbjct 29 VPIDDSELDAITREIDIIKPEVRKLLSSKGDDETSKRKVLLIQSLLSLGLAFHFENEIKD 88
Query 120 TCKDGF 125
+D F
Sbjct 89 ILEDAF 94
Lambda K H
0.325 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2059772308
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40