bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7055_orf3
Length=62
Score E
Sequences producing significant alignments: (Bits) Value
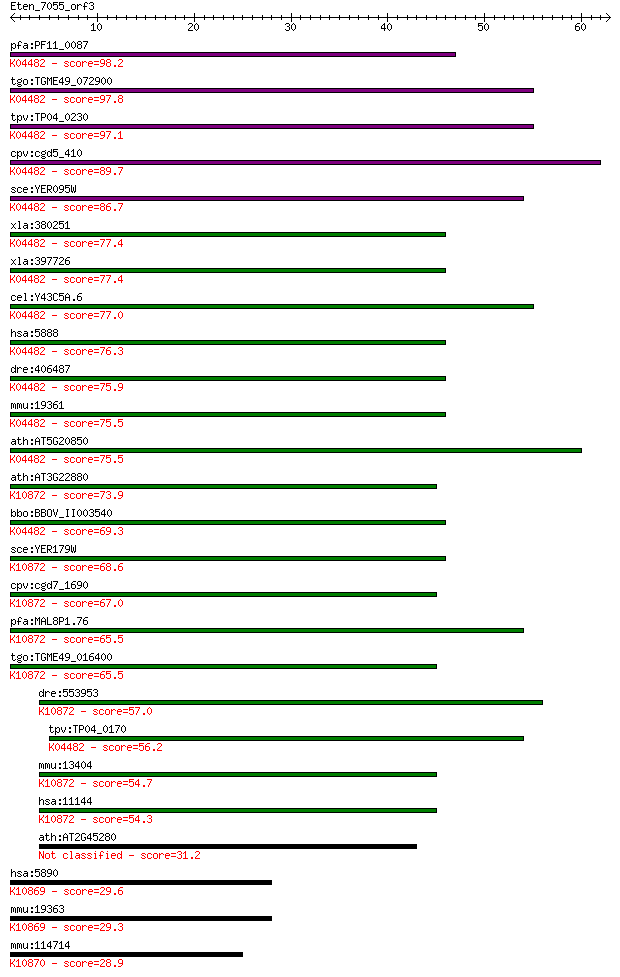
pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein ... 98.2 6e-21
tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8); K... 97.8 8e-21
tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair prot... 97.1 1e-20
cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51 89.7 2e-18
sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51 86.7 2e-17
xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,... 77.4 1e-14
xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad... 77.4 1e-14
cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-... 77.0 1e-14
hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA... 76.3 2e-14
dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA h... 75.9 3e-14
mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cer... 75.5 3e-14
ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-depende... 75.5 4e-14
ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1); AT... 73.9 1e-13
bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair ... 69.3 3e-12
sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination pr... 68.6 5e-12
cpv:cgd7_1690 meiotic recombination protein DMC1-like protein ... 67.0 2e-11
pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;... 65.5 4e-11
tgo:TGME49_016400 meiotic recombination protein DMC1-like prot... 65.5 4e-11
dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor... 57.0 1e-08
tpv:TP04_0170 meiotic recombination protein DMC1; K04482 DNA r... 56.2 3e-08
mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage sup... 54.7 7e-08
hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473, d... 54.3 9e-08
ath:AT2G45280 ATRAD51C; ATP binding / damaged DNA binding / pr... 31.2 0.76
hsa:5890 RAD51L1, MGC34245, R51H2, RAD51B, REC2, hREC2; RAD51-... 29.6 2.3
mmu:19363 Rad51l1, AI553500, R51H2, Rad51b, mREC2; RAD51-like ... 29.3 3.2
mmu:114714 Rad51c, R51H3, Rad51l2; RAD51 homolog c (S. cerevis... 28.9 4.4
> pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein
RAD51
Length=350
Score = 98.2 bits (243), Expect = 6e-21, Method: Composition-based stats.
Identities = 40/46 (86%), Positives = 44/46 (95%), Gaps = 0/46 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNC 46
GGEGKCLWIDTEGTFRPERIVAIA+R+GL P DCL+NIAYA+AYNC
Sbjct 162 GGEGKCLWIDTEGTFRPERIVAIAKRYGLHPTDCLNNIAYAKAYNC 207
> tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8);
K04482 DNA repair protein RAD51
Length=354
Score = 97.8 bits (242), Expect = 8e-21, Method: Composition-based stats.
Identities = 42/54 (77%), Positives = 48/54 (88%), Gaps = 0/54 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLLL 54
GGEGKCLWIDTEGTFRPERIV+IA+RFGL+ DCLDN+AYARAYNC + LL+
Sbjct 165 GGEGKCLWIDTEGTFRPERIVSIAKRFGLNANDCLDNVAYARAYNCDHQMELLM 218
> tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair protein
RAD51
Length=343
Score = 97.1 bits (240), Expect = 1e-20, Method: Composition-based stats.
Identities = 41/54 (75%), Positives = 48/54 (88%), Gaps = 0/54 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLLL 54
GGEGKCLW+D+EGTFRPERIV+IA+RFGL P+DCLDN+AYARAYN L LL+
Sbjct 155 GGEGKCLWVDSEGTFRPERIVSIAKRFGLSPSDCLDNVAYARAYNTDHQLELLV 208
> cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51
Length=347
Score = 89.7 bits (221), Expect = 2e-18, Method: Composition-based stats.
Identities = 41/61 (67%), Positives = 47/61 (77%), Gaps = 0/61 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLLLFGVSVS 60
GGEGKCLWIDTEGTFRPERIV IA RF L+ +DCLDNIAYAR +N + LL V++
Sbjct 158 GGEGKCLWIDTEGTFRPERIVQIADRFNLNASDCLDNIAYARGFNTEHQMDLLQSAVAMM 217
Query 61 T 61
T
Sbjct 218 T 218
> sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51
Length=400
Score = 86.7 bits (213), Expect = 2e-17, Method: Composition-based stats.
Identities = 39/53 (73%), Positives = 45/53 (84%), Gaps = 0/53 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLL 53
GGEGKCL+IDTEGTFRP R+V+IA+RFGLDP D L+N+AYARAYN L LL
Sbjct 210 GGEGKCLYIDTEGTFRPVRLVSIAQRFGLDPDDALNNVAYARAYNADHQLRLL 262
> xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,
reca, xrad51; RAD51 homolog (RecA homolog); K04482 DNA repair
protein RAD51
Length=336
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 31/45 (68%), Positives = 40/45 (88%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGK ++IDTEGTFRPER++A+A R+GL +D LDN+AYARA+N
Sbjct 149 GGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFN 193
> xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad51;
RAD51 homolog (RecA homolog); K04482 DNA repair protein
RAD51
Length=336
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 31/45 (68%), Positives = 40/45 (88%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGK ++IDTEGTFRPER++A+A R+GL +D LDN+AYARA+N
Sbjct 149 GGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFN 193
> cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-related
family member (rad-51); K04482 DNA repair protein
RAD51
Length=395
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 33/54 (61%), Positives = 43/54 (79%), Gaps = 0/54 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLLL 54
GGEGKC++IDT TFRPERI+AIA+R+ +D A L+NIA ARAYN L+ L++
Sbjct 206 GGEGKCMYIDTNATFRPERIIAIAQRYNMDSAHVLENIAVARAYNSEHLMALII 259
> hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA;
RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae); K04482
DNA repair protein RAD51
Length=340
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 31/45 (68%), Positives = 40/45 (88%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGK ++IDTEGTFRPER++A+A R+GL +D LDN+AYARA+N
Sbjct 153 GGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFN 197
> dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA
homolog, E. coli) (S. cerevisiae); K04482 DNA repair protein
RAD51
Length=338
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 31/45 (68%), Positives = 40/45 (88%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGK ++IDTEGTFRPER++A+A R+GL +D LDN+AYARA+N
Sbjct 151 GGEGKAMYIDTEGTFRPERLLAVAERYGLVGSDVLDNVAYARAFN 195
> mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cerevisiae);
K04482 DNA repair protein RAD51
Length=339
Score = 75.5 bits (184), Expect = 3e-14, Method: Composition-based stats.
Identities = 30/45 (66%), Positives = 39/45 (86%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGK ++IDTEGTFRPER++A+A R+GL +D LDN+AYAR +N
Sbjct 152 GGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARGFN 196
> ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-dependent
ATPase/ damaged DNA binding / nucleoside-triphosphatase/
nucleotide binding / protein binding / sequence-specific DNA
binding; K04482 DNA repair protein RAD51
Length=342
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 35/59 (59%), Positives = 44/59 (74%), Gaps = 0/59 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLLLFGVSV 59
GGEGK ++ID EGTFRP+R++ IA RFGL+ AD L+N+AYARAYN LLL S+
Sbjct 155 GGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAYNTDHQSRLLLEAASM 213
> ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1);
ATP binding / DNA binding / DNA-dependent ATPase/ damaged DNA
binding / nucleoside-triphosphatase/ nucleotide binding /
protein binding; K10872 meiotic recombination protein DMC1
Length=344
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 32/44 (72%), Positives = 35/44 (79%), Gaps = 0/44 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAY 44
GG GK +IDTEGTFRP+RIV IA RFG+DP LDNI YARAY
Sbjct 158 GGNGKVAYIDTEGTFRPDRIVPIAERFGMDPGAVLDNIIYARAY 201
> bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair
protein RAD51
Length=346
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 28/45 (62%), Positives = 34/45 (75%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGKCLWIDT+ +FRPER+ IA RFGL A+C+ NI Y + N
Sbjct 155 GGEGKCLWIDTQNSFRPERLGPIANRFGLSHAECVANIVYVKVSN 199
> sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination
protein DMC1
Length=334
Score = 68.6 bits (166), Expect = 5e-12, Method: Composition-based stats.
Identities = 30/45 (66%), Positives = 34/45 (75%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGK +IDTEGTFRPERI IA + LDP CL N++YARA N
Sbjct 146 GGEGKVAYIDTEGTFRPERIKQIAEGYELDPESCLANVSYARALN 190
> cpv:cgd7_1690 meiotic recombination protein DMC1-like protein
; K10872 meiotic recombination protein DMC1
Length=342
Score = 67.0 bits (162), Expect = 2e-11, Method: Composition-based stats.
Identities = 31/44 (70%), Positives = 33/44 (75%), Gaps = 0/44 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAY 44
GG GK +IDTEGTFRPERIV IA RFG+ LDNI YARAY
Sbjct 155 GGNGKVCFIDTEGTFRPERIVKIAERFGVQGDVALDNIMYARAY 198
> pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;
K10872 meiotic recombination protein DMC1
Length=347
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 31/53 (58%), Positives = 36/53 (67%), Gaps = 0/53 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLL 53
GG GK +IDTEGTFRPE++ IA R+GLD LDNI YARA+ L LL
Sbjct 160 GGNGKVCYIDTEGTFRPEKVCKIAERYGLDGEAVLDNILYARAFTHEHLYQLL 212
> tgo:TGME49_016400 meiotic recombination protein DMC1-like protein,
putative (EC:2.7.11.1); K10872 meiotic recombination
protein DMC1
Length=349
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 30/44 (68%), Positives = 33/44 (75%), Gaps = 0/44 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAY 44
GG GK +IDTEGTFRPE+I IA RFGLD LDNI YARA+
Sbjct 162 GGCGKVCYIDTEGTFRPEKIQGIAERFGLDGDGVLDNIMYARAF 205
> dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=342
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 28/52 (53%), Positives = 34/52 (65%), Gaps = 0/52 (0%)
Query 4 GKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLLLF 55
GK ++IDTE TFRPER+ IA RF +D LDN+ YARAY + LL F
Sbjct 156 GKVIFIDTENTFRPERLKDIADRFNVDHEAVLDNVLYARAYTSEHQMELLDF 207
> tpv:TP04_0170 meiotic recombination protein DMC1; K04482 DNA
repair protein RAD51
Length=346
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 27/49 (55%), Positives = 32/49 (65%), Gaps = 0/49 (0%)
Query 5 KCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLL 53
K +IDTE TFRPE+I I RF LDP LDNI Y++AY LL L+
Sbjct 163 KVCYIDTENTFRPEKIEKICERFDLDPMITLDNILYSKAYTNEHLLQLI 211
> mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=340
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 24/41 (58%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 4 GKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAY 44
GK ++IDTE TFRP+R+ IA RF +D LDN+ YARAY
Sbjct 154 GKIIFIDTENTFRPDRLRDIADRFNVDHEAVLDNVLYARAY 194
> hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473,
dJ199H16.1; DMC1 dosage suppressor of mck1 homolog, meiosis-specific
homologous recombination (yeast); K10872 meiotic recombination
protein DMC1
Length=340
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 24/41 (58%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 4 GKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAY 44
GK ++IDTE TFRP+R+ IA RF +D LDN+ YARAY
Sbjct 154 GKIIFIDTENTFRPDRLRDIADRFNVDHDAVLDNVLYARAY 194
> ath:AT2G45280 ATRAD51C; ATP binding / damaged DNA binding /
protein binding / recombinase/ single-stranded DNA binding
Length=363
Score = 31.2 bits (69), Expect = 0.76, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 26/59 (44%), Gaps = 20/59 (33%)
Query 4 GKCLWIDTEGTFRPERIVAIAR--------------------RFGLDPADCLDNIAYAR 42
GK ++IDTEG+F ER + IA + + P D L+NI Y R
Sbjct 158 GKAIYIDTEGSFMVERALQIAEACVEDMEEYTGYMHKHFQANQVQMKPEDILENIFYFR 216
> hsa:5890 RAD51L1, MGC34245, R51H2, RAD51B, REC2, hREC2; RAD51-like
1 (S. cerevisiae); K10869 RAD51-like protein 1
Length=350
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 18/28 (64%), Gaps = 1/28 (3%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIAR-RF 27
G EG ++IDTE F ER+V IA RF
Sbjct 133 GLEGAVVYIDTESAFSAERLVEIAESRF 160
> mmu:19363 Rad51l1, AI553500, R51H2, Rad51b, mREC2; RAD51-like
1 (S. cerevisiae); K10869 RAD51-like protein 1
Length=350
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 18/28 (64%), Gaps = 1/28 (3%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIAR-RF 27
G EG ++IDTE F ER+V IA RF
Sbjct 133 GLEGAVVYIDTESAFTAERLVEIAESRF 160
> mmu:114714 Rad51c, R51H3, Rad51l2; RAD51 homolog c (S. cerevisiae);
K10870 RAD51-like protein 2
Length=366
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIA 24
G G+ ++IDTEG+F +R+V++A
Sbjct 141 GVAGEAVFIDTEGSFMVDRVVSLA 164
Lambda K H
0.326 0.145 0.467
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2060478756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40