bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7048_orf1
Length=115
Score E
Sequences producing significant alignments: (Bits) Value
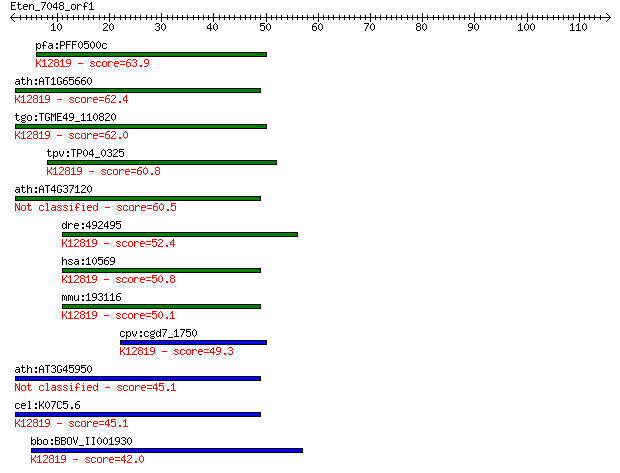
pfa:PFF0500c step II splicing factor, putative; K12819 pre-mRN... 63.9 1e-10
ath:AT1G65660 SMP1; SMP1 (SWELLMAP 1); nucleic acid binding / ... 62.4 4e-10
tgo:TGME49_110820 step II splicing factor SLU7, putative ; K12... 62.0 5e-10
tpv:TP04_0325 step II splicing factor; K12819 pre-mRNA-process... 60.8 1e-09
ath:AT4G37120 SMP2; SMP2; single-stranded RNA binding 60.5 1e-09
dre:492495 slu7, wu:fc94e11, zgc:103640; SLU7 splicing factor ... 52.4 3e-07
hsa:10569 SLU7, 9G8, MGC9280, hSlu7; SLU7 splicing factor homo... 50.8 9e-07
mmu:193116 Slu7, AU018913, D11Ertd730e, D3Bwg0878e, MGC31026; ... 50.1 2e-06
cpv:cgd7_1750 step II splicing factor SLU7 ; K12819 pre-mRNA-p... 49.3 3e-06
ath:AT3G45950 splicing factor-related 45.1 6e-05
cel:K07C5.6 hypothetical protein; K12819 pre-mRNA-processing f... 45.1 6e-05
bbo:BBOV_II001930 18.m06149; mRNA processing-related protein; ... 42.0 5e-04
> pfa:PFF0500c step II splicing factor, putative; K12819 pre-mRNA-processing
factor SLU7
Length=444
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 25/44 (56%), Positives = 32/44 (72%), Gaps = 0/44 (0%)
Query 6 QAAAAAAAAVDAAGKEINPHIPQYISRAPWYLNQQQPGLQHQRF 49
+ A A D G +INPH+PQYI +APWYLNQ +PGL+HQR+
Sbjct 21 RKAGKVEALKDEEGNDINPHMPQYIIKAPWYLNQTKPGLKHQRY 64
> ath:AT1G65660 SMP1; SMP1 (SWELLMAP 1); nucleic acid binding
/ single-stranded RNA binding; K12819 pre-mRNA-processing factor
SLU7
Length=535
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 28/47 (59%), Positives = 35/47 (74%), Gaps = 0/47 (0%)
Query 2 VQTPQAAAAAAAAVDAAGKEINPHIPQYISRAPWYLNQQQPGLQHQR 48
++ + A A A VD GKEINPHIPQY+S APWYLN ++P L+HQR
Sbjct 20 LEEARKAGLAPAEVDEDGKEINPHIPQYMSSAPWYLNSEKPSLKHQR 66
> tgo:TGME49_110820 step II splicing factor SLU7, putative ; K12819
pre-mRNA-processing factor SLU7
Length=544
Score = 62.0 bits (149), Expect = 5e-10, Method: Composition-based stats.
Identities = 29/48 (60%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 2 VQTPQAAAAAAAAVDAAGKEINPHIPQYISRAPWYLNQQQPGLQHQRF 49
++ + A A D G INPHIPQYIS+APWYLNQQ+PGL+HQRF
Sbjct 20 LEEARKAGTAEPEKDEEGNAINPHIPQYISKAPWYLNQQKPGLKHQRF 67
> tpv:TP04_0325 step II splicing factor; K12819 pre-mRNA-processing
factor SLU7
Length=387
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 26/44 (59%), Positives = 31/44 (70%), Gaps = 0/44 (0%)
Query 8 AAAAAAAVDAAGKEINPHIPQYISRAPWYLNQQQPGLQHQRFAE 51
A A D G +INPHIPQYIS+APWYL+Q +P L+HQR E
Sbjct 24 AGTVPALKDELGNDINPHIPQYISKAPWYLDQGEPSLRHQRVNE 67
> ath:AT4G37120 SMP2; SMP2; single-stranded RNA binding
Length=536
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 26/47 (55%), Positives = 35/47 (74%), Gaps = 0/47 (0%)
Query 2 VQTPQAAAAAAAAVDAAGKEINPHIPQYISRAPWYLNQQQPGLQHQR 48
++ + A A A VD GKEINPHIP+Y+S+APWYL +QP L+HQ+
Sbjct 20 LEEARKAGLAPAEVDEDGKEINPHIPEYMSKAPWYLKSEQPSLKHQK 66
> dre:492495 slu7, wu:fc94e11, zgc:103640; SLU7 splicing factor
homolog (S. cerevisiae); K12819 pre-mRNA-processing factor
SLU7
Length=571
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 26/46 (56%), Positives = 33/46 (71%), Gaps = 1/46 (2%)
Query 11 AAAAVDAAGKEINPHIPQYISRAPWYLN-QQQPGLQHQRFAERSSS 55
A A VD GK+INPHIPQYIS PWY++ ++P L+HQR E + S
Sbjct 41 APAEVDEEGKDINPHIPQYISSVPWYIDPSKRPTLKHQRPQEENQS 86
> hsa:10569 SLU7, 9G8, MGC9280, hSlu7; SLU7 splicing factor homolog
(S. cerevisiae); K12819 pre-mRNA-processing factor SLU7
Length=586
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 24/39 (61%), Positives = 30/39 (76%), Gaps = 1/39 (2%)
Query 11 AAAAVDAAGKEINPHIPQYISRAPWYLN-QQQPGLQHQR 48
A A VD GK+INPHIPQYIS PWY++ ++P L+HQR
Sbjct 48 APAEVDEEGKDINPHIPQYISSVPWYIDPSKRPTLKHQR 86
> mmu:193116 Slu7, AU018913, D11Ertd730e, D3Bwg0878e, MGC31026;
SLU7 splicing factor homolog (S. cerevisiae); K12819 pre-mRNA-processing
factor SLU7
Length=585
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 24/39 (61%), Positives = 30/39 (76%), Gaps = 1/39 (2%)
Query 11 AAAAVDAAGKEINPHIPQYISRAPWYLN-QQQPGLQHQR 48
A A VD GK+INPHIPQYIS PWY++ ++P L+HQR
Sbjct 48 APAEVDEEGKDINPHIPQYISSVPWYIDPSKRPTLKHQR 86
> cpv:cgd7_1750 step II splicing factor SLU7 ; K12819 pre-mRNA-processing
factor SLU7
Length=405
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 18/28 (64%), Positives = 25/28 (89%), Gaps = 0/28 (0%)
Query 22 INPHIPQYISRAPWYLNQQQPGLQHQRF 49
+NPHIPQ+I++APWY+NQQ+ L+HQR
Sbjct 30 LNPHIPQFIAKAPWYINQQESSLEHQRL 57
> ath:AT3G45950 splicing factor-related
Length=385
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 2 VQTPQAAAAAAAAVDAAGKEINPHIPQYISRAPWYLNQQQPGLQHQR 48
++ + A A A VD GKEIN HIP+Y++ P Y ++P L+HQ+
Sbjct 20 LEEARKAGLAPAEVDEGGKEINLHIPKYLTIPPLYAKSEKPSLKHQK 66
> cel:K07C5.6 hypothetical protein; K12819 pre-mRNA-processing
factor SLU7
Length=647
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 22/48 (45%), Positives = 32/48 (66%), Gaps = 1/48 (2%)
Query 2 VQTPQAAAAAAAAVDA-AGKEINPHIPQYISRAPWYLNQQQPGLQHQR 48
++ + A A A VD G++INPHIP +IS+ PWY+ + P L+HQR
Sbjct 35 LEEDRKAGTAPAMVDVQTGRDINPHIPMFISQNPWYVPSEGPTLKHQR 82
> bbo:BBOV_II001930 18.m06149; mRNA processing-related protein;
K12819 pre-mRNA-processing factor SLU7
Length=388
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 1/52 (1%)
Query 5 PQAAAAAAAAVDAAGKEINPHIPQYISRAPWYLNQQQPGLQHQRFAERSSSS 56
P A+ E NP+IP++I++APWYLN GL HQR + S+
Sbjct 25 PSVEGASTVEKVVTKDETNPYIPRFIAKAPWYLNATS-GLSHQRVQQHQKSA 75
Lambda K H
0.312 0.124 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2042676840
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40