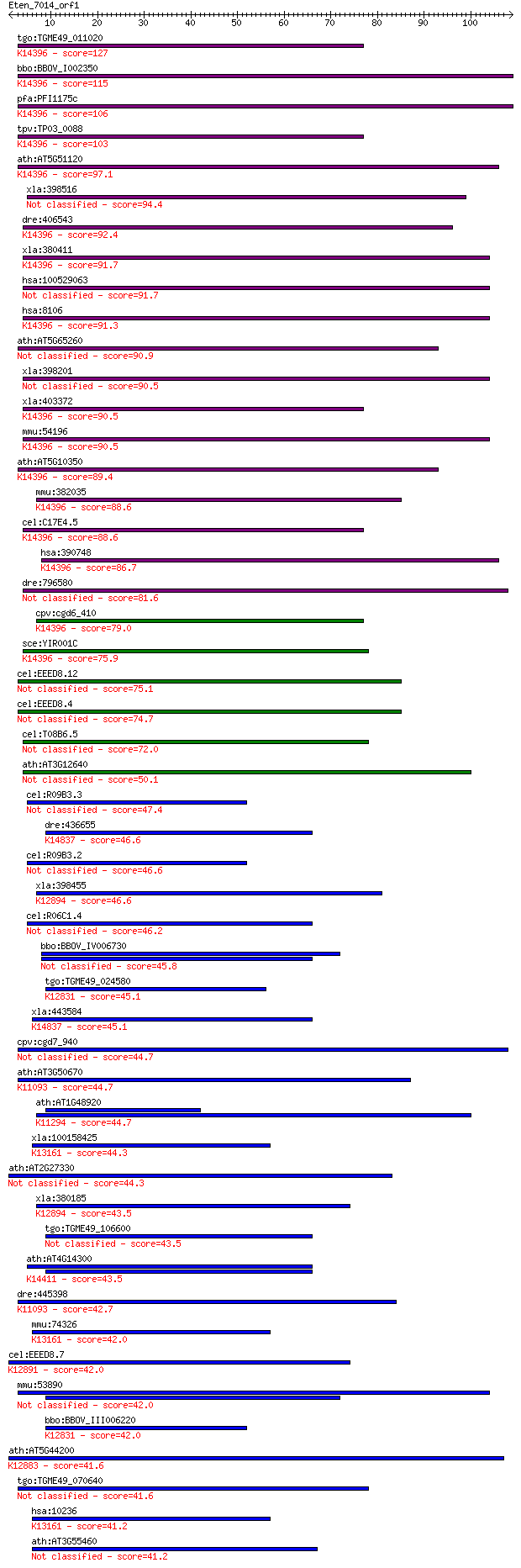
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7014_orf1
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_011020 RNA-binding protein, putative ; K14396 polya... 127 1e-29
bbo:BBOV_I002350 19.m02195; RNA binding protein; K14396 polyad... 115 3e-26
pfa:PFI1175c RNA binding protein, putative; K14396 polyadenyla... 106 2e-23
tpv:TP03_0088 hypothetical protein; K14396 polyadenylate-bindi... 103 1e-22
ath:AT5G51120 PABN1; PABN1 (POLYADENYLATE-BINDING PROTEIN 1); ... 97.1 1e-20
xla:398516 pabpn1l-a, ePABP2, epabp2-a; poly(A) binding protei... 94.4 8e-20
dre:406543 pabpn1, MGC171884, MGC173800, MGC92452, fb19c03, wu... 92.4 3e-19
xla:380411 pabpn1-b, MGC53742, opmd, pab2, pabp2, pabpn1; poly... 91.7 5e-19
hsa:100529063 BCL2L2-PABPN1; BCL2L2-PABPN1 readthrough 91.7 5e-19
hsa:8106 PABPN1, OPMD, PAB2, PABII, PABP-2, PABP2; poly(A) bin... 91.3 6e-19
ath:AT5G65260 polyadenylate-binding protein family protein / P... 90.9 8e-19
xla:398201 pabpn1-a, opmd, pab2, pabp2, pabpii; poly(A) bindin... 90.5 1e-18
xla:403372 pabpn1l-b, ePABP2, epabp2-b, pabpn2; poly(A) bindin... 90.5 1e-18
mmu:54196 Pabpn1, PAB2, Pabp3, mPABII; poly(A) binding protein... 90.5 1e-18
ath:AT5G10350 polyadenylate-binding protein family protein / P... 89.4 3e-18
mmu:382035 Pabpn1l, Gm1108, Pabpnl1, ePABP2; poly(A)binding pr... 88.6 4e-18
cel:C17E4.5 pabp-2; PolyA Binding Protein (nuclear) family mem... 88.6 4e-18
hsa:390748 PABPN1L, PABPNL1, ePABP2; poly(A) binding protein, ... 86.7 1e-17
dre:796580 Polyadenylate-binding protein 2-like 81.6 6e-16
cpv:cgd6_410 Sgn1p-like RRM domain containing protein ; K14396... 79.0 4e-15
sce:YIR001C SGN1, RBP1, RBP29; Cytoplasmic RNA-binding protein... 75.9 3e-14
cel:EEED8.12 hypothetical protein 75.1 5e-14
cel:EEED8.4 hypothetical protein 74.7 7e-14
cel:T08B6.5 hypothetical protein 72.0 4e-13
ath:AT3G12640 RNA binding / nucleic acid binding / nucleotide ... 50.1 2e-06
cel:R09B3.3 hypothetical protein 47.4 1e-05
dre:436655 rbm34, zgc:92062; RNA binding motif protein 34; K14... 46.6 2e-05
cel:R09B3.2 hypothetical protein 46.6 2e-05
xla:398455 hnrnpa0, MGC130809, hnrpa0; heterogeneous nuclear r... 46.6 2e-05
cel:R06C1.4 hypothetical protein 46.2 3e-05
bbo:BBOV_IV006730 23.m05875; single stranded G-strand telomeri... 45.8 3e-05
tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12... 45.1 6e-05
xla:443584 rbm34, MGC115058; RNA binding motif protein 34; K14... 45.1 6e-05
cpv:cgd7_940 splicing factor RRM domain containing protein; T2... 44.7 6e-05
ath:AT3G50670 U1-70K; U1-70K (U1 SMALL NUCLEAR RIBONUCLEOPROTE... 44.7 8e-05
ath:AT1G48920 ATNUC-L1; nucleic acid binding / nucleotide bind... 44.7 8e-05
xla:100158425 hnrnpr; heterogeneous nuclear ribonucleoprotein ... 44.3 1e-04
ath:AT2G27330 RNA recognition motif (RRM)-containing protein 44.3 1e-04
xla:380185 hnrpa0, MGC53160; heterogeneous nuclear ribonucleop... 43.5 2e-04
tgo:TGME49_106600 RNA recognition motif domain-containing protein 43.5 2e-04
ath:AT4G14300 heterogeneous nuclear ribonucleoprotein, putativ... 43.5 2e-04
dre:445398 snrnp70, MGC136450, snrp70, wu:fc20b04, zgc:136450;... 42.7 3e-04
mmu:74326 Hnrnpr, 2610003J05Rik, Hnrpr; heterogeneous nuclear ... 42.0 4e-04
cel:EEED8.7 rsp-4; SR Protein (splicing factor) family member ... 42.0 4e-04
mmu:53890 Sart3, AU045857, mKIAA0156; squamous cell carcinoma ... 42.0 5e-04
bbo:BBOV_III006220 17.m07552; RNA recognition motif domaining ... 42.0 5e-04
ath:AT5G44200 CBP20; CBP20 (CAP-BINDING PROTEIN 20); RNA bindi... 41.6 5e-04
tgo:TGME49_070640 RNA binding motif-containing protein 41.6 6e-04
hsa:10236 HNRNPR, FLJ25714, HNRPR, hnRNP_R, hnRNP-R; heterogen... 41.2 8e-04
ath:AT3G55460 SCL30; SCL30; RNA binding / nucleic acid binding... 41.2 9e-04
> tgo:TGME49_011020 RNA-binding protein, putative ; K14396 polyadenylate-binding
protein 2
Length=268
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 57/74 (77%), Positives = 65/74 (87%), Gaps = 0/74 (0%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
S+TPA+LQEHFK CG INRITIMVDK+TGHPKGYAYIEF SE A Q+A+LLSD+ K RQ
Sbjct 149 SSTPAELQEHFKSCGTINRITIMVDKYTGHPKGYAYIEFNSEAAVQNAILLSDTVFKQRQ 208
Query 63 IKVVSKRKNIPGMS 76
IKVV+KRKNIPG +
Sbjct 209 IKVVAKRKNIPGFA 222
> bbo:BBOV_I002350 19.m02195; RNA binding protein; K14396 polyadenylate-binding
protein 2
Length=163
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 64/106 (60%), Positives = 71/106 (66%), Gaps = 11/106 (10%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
S P DLQE FK G INRITIMVDK+TG PKGYAYIEF SE A +AV+L+D K+R
Sbjct 52 STKPHDLQEFFKASGQINRITIMVDKWTGKPKGYAYIEFASEDAVNNAVMLNDCLFKERI 111
Query 63 IKVVSKRKNIPGMSRGRGGGWRPRGAVPFRGRGRGGGSFRPTYRGR 108
IKV+ KRKNIPG +R R A P RGRGRG FRP GR
Sbjct 112 IKVIPKRKNIPGYNRKR--------AQPSRGRGRG---FRPMRGGR 146
> pfa:PFI1175c RNA binding protein, putative; K14396 polyadenylate-binding
protein 2
Length=202
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 59/109 (54%), Positives = 72/109 (66%), Gaps = 4/109 (3%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
S P +LQ F CG INR+TI+V+K TGH KGYAYIEF + + A+ LS+S K RQ
Sbjct 81 STQPEELQSLFSECGLINRVTILVNKNTGHSKGYAYIEFADASSVRTALSLSESFFKKRQ 140
Query 63 IKVVSKRKNIPGMSRGRGGGWRPRG---AVPFRGRGRGGGSFRPTYRGR 108
IKV SKR+NIPG +R + +R RG A+ RGR R SFRP YRGR
Sbjct 141 IKVCSKRRNIPGFNRPKISPFRGRGMKSALGVRGRLR-QPSFRPFYRGR 188
> tpv:TP03_0088 hypothetical protein; K14396 polyadenylate-binding
protein 2
Length=151
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 46/74 (62%), Positives = 58/74 (78%), Gaps = 0/74 (0%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
S P +LQE FK G INRITIMVDK+TGHPKGYAY+EF +E A +A++L++S K+R
Sbjct 52 STKPQELQEFFKSSGQINRITIMVDKYTGHPKGYAYVEFSNEDAVNNAIMLNESLFKERI 111
Query 63 IKVVSKRKNIPGMS 76
IKV+ KRKNIPG +
Sbjct 112 IKVIPKRKNIPGFN 125
> ath:AT5G51120 PABN1; PABN1 (POLYADENYLATE-BINDING PROTEIN 1);
RNA binding / poly(A) binding / protein binding; K14396 polyadenylate-binding
protein 2
Length=227
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 51/106 (48%), Positives = 71/106 (66%), Gaps = 11/106 (10%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
+ TP ++Q+HF+ CG +NR+TI+ DKF G PKG+AY+EF A Q++++L++S L RQ
Sbjct 113 ACTPEEVQQHFQSCGTVNRVTILTDKF-GQPKGFAYVEFVEVEAVQNSLILNESELHGRQ 171
Query 63 IKVVSKRKNIPGMS--RGRGGGWRP-RGAVPFRGRGRGGGSFRPTY 105
IKV +KR N+PGM RGRG +RP RG +P G F P Y
Sbjct 172 IKVSAKRTNVPGMRQFRGRGRPFRPMRGFMP-------GVPFYPPY 210
> xla:398516 pabpn1l-a, ePABP2, epabp2-a; poly(A) binding protein,
nuclear 1-like (cytoplasmic)
Length=218
Score = 94.4 bits (233), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 52/109 (47%), Positives = 63/109 (57%), Gaps = 16/109 (14%)
Query 5 TPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIK 64
T DL+ HF CG INRITI+ DKF+GHPKGYAYIEF + AV + ++ + R IK
Sbjct 105 TAQDLEAHFSSCGSINRITILCDKFSGHPKGYAYIEFAERNSVDAAVTMDETVFRGRTIK 164
Query 65 VVSKRKNIPGMSRGRGG---------------GWRPRGAVPFRGRGRGG 98
V+ KR N+PG+S G G RPRG PFRG GR G
Sbjct 165 VLPKRTNMPGISSTDRGGFRGRPRGNRGNYQRGQRPRGR-PFRGCGRPG 212
> dre:406543 pabpn1, MGC171884, MGC173800, MGC92452, fb19c03,
wu:fb17e03, wu:fb18c06, wu:fb19c03, zgc:85979, zgc:92452; poly(A)
binding protein, nuclear 1; K14396 polyadenylate-binding
protein 2
Length=232
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 47/92 (51%), Positives = 61/92 (66%), Gaps = 4/92 (4%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
AT +L+ HF GCG +NR+TI+ DKFTGHPKG+AYIEF + + + A+ L +S + RQI
Sbjct 109 ATAEELEAHFHGCGSVNRVTILCDKFTGHPKGFAYIEFADKESVRTAMALDESLFRGRQI 168
Query 64 KVVSKRKNIPGMSRGRGGGWRPRGAVPFRGRG 95
KV +KR N PG+S G R R FR RG
Sbjct 169 KVGAKRTNRPGISTTDRGFPRAR----FRSRG 196
> xla:380411 pabpn1-b, MGC53742, opmd, pab2, pabp2, pabpn1; poly(A)
binding protein, nuclear 1; K14396 polyadenylate-binding
protein 2
Length=295
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 53/123 (43%), Positives = 69/123 (56%), Gaps = 24/123 (19%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
AT +L+ HF GCG +NR+TI+ DKFTGHPKG+AYIEF + + + ++ L +S + RQI
Sbjct 173 ATAEELEAHFHGCGSVNRVTILCDKFTGHPKGFAYIEFSDKESVRTSLALDESLFRGRQI 232
Query 64 KVVSKRKNIPGMS---RG--------------------RGGGWRPRGAVPFRGRGRGGGS 100
KVV KR N PG+S RG G RPRG V +RGR R
Sbjct 233 KVVPKRTNRPGISTTDRGFPRARYRARASSYSSRSRFYSGYTPRPRGRV-YRGRARATSW 291
Query 101 FRP 103
+ P
Sbjct 292 YTP 294
> hsa:100529063 BCL2L2-PABPN1; BCL2L2-PABPN1 readthrough
Length=333
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 50/124 (40%), Positives = 69/124 (55%), Gaps = 25/124 (20%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
AT +L+ HF GCG +NR+TI+ DKF+GHPKG+AYIEF + + + ++ L +S + RQI
Sbjct 210 ATAEELEAHFHGCGSVNRVTILCDKFSGHPKGFAYIEFSDKESVRTSLALDESLFRGRQI 269
Query 64 KVVSKRKNIPGMS-----------RGRGGGW-------------RPRGAVPFRGRGRGGG 99
KV+ KR N PG+S R R + RPRG V +RGR R
Sbjct 270 KVIPKRTNRPGISTTDRGFPRARYRARTTNYNSSRSRFYSGFNSRPRGRV-YRGRARATS 328
Query 100 SFRP 103
+ P
Sbjct 329 WYSP 332
> hsa:8106 PABPN1, OPMD, PAB2, PABII, PABP-2, PABP2; poly(A) binding
protein, nuclear 1; K14396 polyadenylate-binding protein
2
Length=306
Score = 91.3 bits (225), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 50/124 (40%), Positives = 69/124 (55%), Gaps = 25/124 (20%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
AT +L+ HF GCG +NR+TI+ DKF+GHPKG+AYIEF + + + ++ L +S + RQI
Sbjct 183 ATAEELEAHFHGCGSVNRVTILCDKFSGHPKGFAYIEFSDKESVRTSLALDESLFRGRQI 242
Query 64 KVVSKRKNIPGMS-----------RGRGGGW-------------RPRGAVPFRGRGRGGG 99
KV+ KR N PG+S R R + RPRG V +RGR R
Sbjct 243 KVIPKRTNRPGISTTDRGFPRARYRARTTNYNSSRSRFYSGFNSRPRGRV-YRGRARATS 301
Query 100 SFRP 103
+ P
Sbjct 302 WYSP 305
> ath:AT5G65260 polyadenylate-binding protein family protein /
PABP family protein
Length=220
Score = 90.9 bits (224), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 41/90 (45%), Positives = 61/90 (67%), Gaps = 1/90 (1%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
+ TP ++Q+HF+ CG ++R+TI+ DKF G PKG+AY+EF A Q A+ L++S L RQ
Sbjct 102 ACTPEEVQQHFQTCGTVHRVTILTDKF-GQPKGFAYVEFVEVEAVQEALQLNESELHGRQ 160
Query 63 IKVVSKRKNIPGMSRGRGGGWRPRGAVPFR 92
+KV+ KR N+PG+ + RG + P FR
Sbjct 161 LKVLQKRTNVPGLKQFRGRRFNPYMGYRFR 190
> xla:398201 pabpn1-a, opmd, pab2, pabp2, pabpii; poly(A) binding
protein, nuclear 1
Length=296
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 53/123 (43%), Positives = 69/123 (56%), Gaps = 24/123 (19%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
AT +L+ HF GCG +NR+TI+ DKFTGHPKG+AYIEF + + + ++ L +S + RQI
Sbjct 174 ATAEELEAHFHGCGSVNRVTILCDKFTGHPKGFAYIEFCDKESVRTSLALDESLFRGRQI 233
Query 64 KVVSKRKNIPGMS---RG--------------------RGGGWRPRGAVPFRGRGRGGGS 100
KVV KR N PG+S RG G RPRG V +RGR R
Sbjct 234 KVVPKRTNRPGISTTDRGFPRARYRARASSYSSRSRFYSGYTPRPRGRV-YRGRARATSW 292
Query 101 FRP 103
+ P
Sbjct 293 YTP 295
> xla:403372 pabpn1l-b, ePABP2, epabp2-b, pabpn2; poly(A) binding
protein, nuclear 1-like (cytoplasmic); K14396 polyadenylate-binding
protein 2
Length=218
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/73 (53%), Positives = 51/73 (69%), Gaps = 0/73 (0%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
+T DL+ HF CG INRITI+ DKF+GHPKGYAYIEF + AV + ++ + R I
Sbjct 104 STAQDLEAHFSSCGSINRITILCDKFSGHPKGYAYIEFAERNSVDAAVAMDETVFRGRTI 163
Query 64 KVVSKRKNIPGMS 76
KV+ KR N+PG+S
Sbjct 164 KVLPKRTNMPGIS 176
> mmu:54196 Pabpn1, PAB2, Pabp3, mPABII; poly(A) binding protein,
nuclear 1; K14396 polyadenylate-binding protein 2
Length=302
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 49/124 (39%), Positives = 69/124 (55%), Gaps = 25/124 (20%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
AT +L+ HF GCG +NR+TI+ DKF+GHPKG+AYIEF + + + ++ L +S + RQI
Sbjct 179 ATAEELEAHFHGCGSVNRVTILCDKFSGHPKGFAYIEFSDKESVRTSLALDESLFRGRQI 238
Query 64 KVVSKRKNIPGMS-----------RGRGGGW-------------RPRGAVPFRGRGRGGG 99
KV+ KR N PG+S R R + RPRG + +RGR R
Sbjct 239 KVIPKRTNRPGISTTDRGFPRSRYRARTTNYNSSRSRFYSGFNSRPRGRI-YRGRARATS 297
Query 100 SFRP 103
+ P
Sbjct 298 WYSP 301
> ath:AT5G10350 polyadenylate-binding protein family protein /
PABP family protein; K14396 polyadenylate-binding protein 2
Length=217
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 42/90 (46%), Positives = 59/90 (65%), Gaps = 1/90 (1%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
+ TP ++Q HF+ CG +NR+TI++DKF G PKG+AY+EF A Q A+ L++S L RQ
Sbjct 99 ACTPEEVQLHFQTCGTVNRVTILMDKF-GQPKGFAYVEFVEVEAVQEALQLNESELHGRQ 157
Query 63 IKVVSKRKNIPGMSRGRGGGWRPRGAVPFR 92
+KV KR N+PGM + G + P FR
Sbjct 158 LKVSPKRTNVPGMKQYHPGRFNPSMGYRFR 187
> mmu:382035 Pabpn1l, Gm1108, Pabpnl1, ePABP2; poly(A)binding
protein nuclear 1-like; K14396 polyadenylate-binding protein
2
Length=273
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 42/78 (53%), Positives = 55/78 (70%), Gaps = 0/78 (0%)
Query 7 ADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIKVV 66
A+L+ +F CG I+R+TI+ DKF+GHPKGYAYIEF S + + AV L +ST + R IKV+
Sbjct 157 AELEAYFSPCGEIHRVTILCDKFSGHPKGYAYIEFASHRSVKAAVGLDESTFRGRVIKVL 216
Query 67 SKRKNIPGMSRGRGGGWR 84
KR N PG+S GG R
Sbjct 217 PKRTNFPGISSTDRGGLR 234
> cel:C17E4.5 pabp-2; PolyA Binding Protein (nuclear) family member
(pabp-2); K14396 polyadenylate-binding protein 2
Length=205
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 38/73 (52%), Positives = 55/73 (75%), Gaps = 0/73 (0%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
AT ++++HF GCG ++R+TI D+F+GHPKG+AY+EF + Q+A+ ++DS L+ RQI
Sbjct 89 ATAEEIEQHFHGCGSVSRVTIQCDRFSGHPKGFAYVEFTEKEGMQNALAMTDSLLRGRQI 148
Query 64 KVVSKRKNIPGMS 76
KV KR N PGMS
Sbjct 149 KVDPKRTNKPGMS 161
> hsa:390748 PABPN1L, PABPNL1, ePABP2; poly(A) binding protein,
nuclear 1-like (cytoplasmic); K14396 polyadenylate-binding
protein 2
Length=278
Score = 86.7 bits (213), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 50/115 (43%), Positives = 66/115 (57%), Gaps = 18/115 (15%)
Query 8 DLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIKVVS 67
+L+ HF CG ++R+TI+ DKF+GHPKGYAYIEF ++ + Q AV L S + R IKV+
Sbjct 162 ELEAHFSRCGEVHRVTILCDKFSGHPKGYAYIEFATKGSVQAAVELDQSLFRGRVIKVLP 221
Query 68 KRKNIPGMSRGRGGGWR----PRGAVPF-------------RGRGRGGGSFRPTY 105
KR N PG+S GG R RGA PF +G+ R G F P +
Sbjct 222 KRTNFPGISSTDRGGLRGHPGSRGA-PFPHSGLQGRPRLRPQGQNRARGKFSPWF 275
> dre:796580 Polyadenylate-binding protein 2-like
Length=192
Score = 81.6 bits (200), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 46/107 (42%), Positives = 61/107 (57%), Gaps = 11/107 (10%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
AT +L+ +F CG +NR+TI ++FTGHPKG+AYIEF + + A+ L ++ + R I
Sbjct 77 ATADELEMYFNSCGHVNRVTIPYNRFTGHPKGFAYIEFSDRESVRTAMALDETLFRGRVI 136
Query 64 KVVSKRKNIPGMSRGRG---GGWRPRGAVPFRGRGRGGGSFRPTYRG 107
KV KR NIPG S GGW RGRG F P +RG
Sbjct 137 KVSPKRTNIPGFSTTDVYLRGGWS-------RGRGVRASRFNP-FRG 175
> cpv:cgd6_410 Sgn1p-like RRM domain containing protein ; K14396
polyadenylate-binding protein 2
Length=262
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 38/70 (54%), Positives = 48/70 (68%), Gaps = 0/70 (0%)
Query 7 ADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIKVV 66
+LQ+ FK CG INRITIM DK TG PKG+AY+EF A + A+ + + RQIKV
Sbjct 124 TELQDLFKSCGSINRITIMNDKRTGMPKGFAYLEFCEPEAVETALKFDGAMFRGRQIKVS 183
Query 67 SKRKNIPGMS 76
+KRKNIPG +
Sbjct 184 TKRKNIPGYN 193
> sce:YIR001C SGN1, RBP1, RBP29; Cytoplasmic RNA-binding protein,
contains an RNA recognition motif (RRM); may have a role
in mRNA translation, as suggested by genetic interactions with
genes encoding proteins involved in translational initiation;
K14396 polyadenylate-binding protein 2
Length=250
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/74 (48%), Positives = 48/74 (64%), Gaps = 0/74 (0%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
TP +++HFK CG I RIT++ D+ TG PKGY YIEF S + A+ L+ LK ++I
Sbjct 75 VTPEQIEDHFKDCGQIKRITLLYDRNTGTPKGYGYIEFESPAYREKALQLNGGELKGKKI 134
Query 64 KVVSKRKNIPGMSR 77
V KR NIPG +R
Sbjct 135 AVSRKRTNIPGFNR 148
> cel:EEED8.12 hypothetical protein
Length=197
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 37/82 (45%), Positives = 52/82 (63%), Gaps = 0/82 (0%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
++T +++EHFKGCG I R TI DKFT K +AYIEF + ++A++++ S + R
Sbjct 71 NSTIEEVEEHFKGCGHIVRTTIPKDKFTKKQKNFAYIEFDDSSSIENALVMNGSLFRSRP 130
Query 63 IKVVSKRKNIPGMSRGRGGGWR 84
I V +KR NIPGM G G R
Sbjct 131 IVVTAKRTNIPGMGHGVRGSSR 152
> cel:EEED8.4 hypothetical protein
Length=191
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 36/82 (43%), Positives = 52/82 (63%), Gaps = 0/82 (0%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
++T +++EHFKGCG I + TI DKFT K +AYIEF + ++A++++ S + R
Sbjct 65 NSTIEEIEEHFKGCGQIVKTTIPKDKFTKKQKNFAYIEFDDSSSIENALVMNGSLFRSRP 124
Query 63 IKVVSKRKNIPGMSRGRGGGWR 84
I V +KR NIPGM G G R
Sbjct 125 IVVTAKRTNIPGMGHGVRGSSR 146
> cel:T08B6.5 hypothetical protein
Length=177
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 32/74 (43%), Positives = 50/74 (67%), Gaps = 0/74 (0%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
+T ++++EHF CG + R+TI DKFTG+ K +A++EF + Q A++LS + ++R+I
Sbjct 57 STVSEIEEHFAVCGKVARVTIPKDKFTGYQKNFAFVEFQKLESVQLALVLSGTMFRNRKI 116
Query 64 KVVSKRKNIPGMSR 77
V KR N PGM R
Sbjct 117 MVTLKRTNKPGMGR 130
> ath:AT3G12640 RNA binding / nucleic acid binding / nucleotide
binding
Length=638
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 53/108 (49%), Gaps = 13/108 (12%)
Query 4 ATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQI 63
AT L HF G + + I+ D TG P G AYIEF + AA++A+ L ++ R +
Sbjct 491 ATKDSLSRHFNKFGEVLKAFIVTDPATGQPSGSAYIEFTRKEAAENALSLDGTSFMSRIL 550
Query 64 KVV-----SKRKNIPGMSRGRGGGWRPRGAVPFRG---RG----RGGG 99
K+V ++ MS RGG + R FRG RG RGGG
Sbjct 551 KIVKGSNGQNQEAASSMSWSRGGRF-TRAPSYFRGGAVRGRSVVRGGG 597
> cel:R09B3.3 hypothetical protein
Length=85
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 5 TPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAV 51
T DL +F G ++ + I+ D+ TG P+G+A++EF E AAQ AV
Sbjct 16 TEDDLGNYFSQAGNVSNVRIVCDRETGRPRGFAFVEFTEEAAAQRAV 62
> dre:436655 rbm34, zgc:92062; RNA binding motif protein 34; K14837
nucleolar protein 12
Length=411
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 9 LQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIKV 65
LQ HF+ CG I + ++ D+ +G KG+ Y+ F S + A+ L+ STL+ R+I+V
Sbjct 268 LQNHFQECGNIEAVRLVRDRDSGMGKGFGYVLFESPDSVMLALKLNGSTLQQRKIRV 324
> cel:R09B3.2 hypothetical protein
Length=83
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/47 (44%), Positives = 29/47 (61%), Gaps = 0/47 (0%)
Query 5 TPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAV 51
T +L F G IN + I+ D+ TG P+G+A+IEF E +AQ AV
Sbjct 17 TEEELGNFFSSIGQINNVRIVCDRETGRPRGFAFIEFAEEGSAQRAV 63
> xla:398455 hnrnpa0, MGC130809, hnrpa0; heterogeneous nuclear
ribonucleoprotein A0; K12894 heterogeneous nuclear ribonucleoprotein
A0
Length=308
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 3/74 (4%)
Query 7 ADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIKVV 66
+DL EHF GP+ ++ ++ DK TG +G+ ++ F S +A A ++ ++ +++V
Sbjct 117 SDLLEHFSQFGPVEKVEVIADKLTGKKRGFGFVYFNSHDSADKAAVVKFHSVNGHRVEV- 175
Query 67 SKRKNIPGMSRGRG 80
+K +P +G
Sbjct 176 --KKAVPKEELSQG 187
> cel:R06C1.4 hypothetical protein
Length=84
Score = 46.2 bits (108), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 5 TPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAV-LLSDSTLKDRQI 63
T ++ +F G +N + I+ D+ TG P+G+A++EF E AQ AV L+ R +
Sbjct 18 TEEEIGNYFAAVGHVNNVRIVYDRETGRPRGFAFVEFSEEAGAQRAVEQLNGVAFNGRNL 77
Query 64 KV 65
+V
Sbjct 78 RV 79
> bbo:BBOV_IV006730 23.m05875; single stranded G-strand telomeric
DNA-binding protein
Length=196
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 37/65 (56%), Gaps = 2/65 (3%)
Query 8 DLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAV-LLSDSTLKDRQIKVV 66
DL++H K G + R I+ D F G KG +EF EI AQ A+ L+D+ L DR I V
Sbjct 22 DLKDHMKQVGEVIRADIIED-FDGKSKGCGIVEFVDEITAQRAMDELNDTMLFDRPIFVR 80
Query 67 SKRKN 71
R+N
Sbjct 81 EDREN 85
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Query 8 DLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKD-RQIKV 65
DL++ FK C PINR+ I+ + G +G A + E A + D + D R+I V
Sbjct 132 DLKDLFKTCAPINRVDILT-REDGKSRGVAKVYVQCEEDANALISTYDGYVLDGREICV 189
> tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12831
splicing factor 3B subunit 4
Length=576
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 9 LQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSD 55
L E F CGP+ +++ DK TG+ +GY ++EF +E+ A +A+ L +
Sbjct 45 LWELFVQCGPVRTVSVPRDKLTGNHQGYGFVEFRNEVDADYALKLMN 91
> xla:443584 rbm34, MGC115058; RNA binding motif protein 34; K14837
nucleolar protein 12
Length=414
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 5/65 (7%)
Query 6 PADLQE-----HFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKD 60
P D++E HF CG + + I+ D+ TG KG+ Y+ F S A Q A+ L++S L
Sbjct 281 PYDIEEESIRKHFSQCGDVQGVRIIRDQKTGIGKGFGYVLFESADAVQLALKLNNSQLSG 340
Query 61 RQIKV 65
R+I+V
Sbjct 341 RRIRV 345
> cpv:cgd7_940 splicing factor RRM domain containing protein;
T22E16.120 SC35-like splicing factor
Length=286
Score = 44.7 bits (104), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 37/113 (32%), Positives = 56/113 (49%), Gaps = 8/113 (7%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKD-R 61
+P ++ HF+ GP+ + + +D +T P+G+ ++E+ AQ AV D +L D
Sbjct 100 ETSPGRVRHHFERYGPVRDVYLPLDYYTRRPRGFGFVEYMDPRDAQDAVNRLDGSLLDGS 159
Query 62 QIKVV---SKRKNIPGMSR-GRGGGWRPR-GAVPFRGRGRGGGSFRPT--YRG 107
I+VV +RK+ M R R G PR G P R R G + P YRG
Sbjct 160 TIRVVVAHDRRKSPETMRRIQRDSGRGPRMGGPPSRYDHRPSGGYPPEHGYRG 212
> ath:AT3G50670 U1-70K; U1-70K (U1 SMALL NUCLEAR RIBONUCLEOPROTEIN-70K);
RNA binding / nucleic acid binding / nucleotide binding;
K11093 U1 small nuclear ribonucleoprotein 70kDa
Length=427
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 43/85 (50%), Gaps = 8/85 (9%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
++ + ++ F+ GPI R+ ++ D+ T PKGYA+IE+ + A +D
Sbjct 148 ESSESKIKREFESYGPIKRVHLVTDQLTNKPKGYAFIEYMHTRDMKAAYKQADG------ 201
Query 63 IKVVSKRKNIPGMSRGRG-GGWRPR 86
+ + R+ + + RGR WRPR
Sbjct 202 -QKIDGRRVLVDVERGRTVPNWRPR 225
> ath:AT1G48920 ATNUC-L1; nucleic acid binding / nucleotide binding;
K11294 nucleolin
Length=557
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 9 LQEHFKGCGPINRITIMVDKFTGHPKGYAYIEF 41
L+EHF CG I +++ +D+ TG+ KG AY+EF
Sbjct 421 LREHFSSCGEIKNVSVPIDRDTGNSKGIAYLEF 453
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 43/94 (45%), Gaps = 10/94 (10%)
Query 7 ADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIKVV 66
AD++ FK G + + ++ G +G+ ++EF S AQ A+ L R+I++
Sbjct 311 ADVENFFKEAGEVVDVRFSTNRDDGSFRGFGHVEFASSEEAQKALEFHGRPLLGREIRL- 369
Query 67 SKRKNIPGMSRGRGG-GWRPRGAVPFRGRGRGGG 99
+++ RG G RP P G R GG
Sbjct 370 -------DIAQERGERGERP-AFTPQSGNFRSGG 395
> xla:100158425 hnrnpr; heterogeneous nuclear ribonucleoprotein
R; K13161 heterogeneous nuclear ribonucleoprotein R
Length=511
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 33/56 (58%), Gaps = 5/56 (8%)
Query 6 PADLQEH-----FKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDS 56
P DL E F+ GPI + +M+D +G +GYA+I F ++ AAQ AV L D+
Sbjct 72 PRDLFEDELVPLFEKAGPIWDLRLMMDPLSGQNRGYAFITFCNKEAAQEAVKLCDN 127
> ath:AT2G27330 RNA recognition motif (RRM)-containing protein
Length=116
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 44/86 (51%), Gaps = 4/86 (4%)
Query 1 STSATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKD 60
S S+T L + F G + ++ +++DK PKG+AY+ F S+ A+ A+L ++ L D
Sbjct 29 SFSSTEETLTQAFSQYGQVLKVDVIMDKIRCRPKGFAYVTFSSKEEAEKALLELNAQLVD 88
Query 61 RQIKVVSKRK----NIPGMSRGRGGG 82
++ ++ K N P GG
Sbjct 89 GRVVILDTTKAAKHNPPDTKPNHAGG 114
> xla:380185 hnrpa0, MGC53160; heterogeneous nuclear ribonucleoprotein
A0; K12894 heterogeneous nuclear ribonucleoprotein
A0
Length=308
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 38/67 (56%), Gaps = 3/67 (4%)
Query 7 ADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIKVV 66
+DL EHF G + R+ I+ DK +G +G+ ++ F S +A A ++ ++ +++V
Sbjct 117 SDLLEHFSQFGAVERVEIIADKLSGKKRGFGFVYFASHDSADKAAVVKFHSVNGHRVEV- 175
Query 67 SKRKNIP 73
+K +P
Sbjct 176 --KKAVP 180
> tgo:TGME49_106600 RNA recognition motif domain-containing protein
Length=189
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 34/61 (55%), Gaps = 7/61 (11%)
Query 9 LQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGS----EIAAQHAVLLSDSTLKDRQIK 64
L+E F G + T+ VDK G PKGYAY+EF S E+A +H L+ L R +K
Sbjct 23 LREIFGLYGVVTSCTVAVDKVAGIPKGYAYVEFDSVREAELAREH---LNQGMLDGRAMK 79
Query 65 V 65
V
Sbjct 80 V 80
> ath:AT4G14300 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=411
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 34/61 (55%), Gaps = 0/61 (0%)
Query 5 TPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIK 64
T + +++F+ GP+ + IM D+ T P+G+ ++ F SE A + + L +Q++
Sbjct 122 TDEEFRQYFEVYGPVTDVAIMYDQATNRPRGFGFVSFDSEDAVDSVLHKTFHDLSGKQVE 181
Query 65 V 65
V
Sbjct 182 V 182
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query 9 LQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIKV 65
L+EHF G +++ +M DK TG P+G+ ++ F S+ + VL ++ R++ V
Sbjct 22 LREHFTNYGEVSQAIVMRDKLTGRPRGFGFVIF-SDPSVLDRVLQEKHSIDTREVDV 77
> dre:445398 snrnp70, MGC136450, snrp70, wu:fc20b04, zgc:136450;
small nuclear ribonucleoprotein 70 (U1); K11093 U1 small
nuclear ribonucleoprotein 70kDa
Length=495
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 41/82 (50%), Gaps = 8/82 (9%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQ 62
T + L+ F+ GPI RI I+ +K TG P+GYA+IE+ E A +D
Sbjct 113 DTTESKLRREFEVYGPIKRIYIVYNKKTGKPRGYAFIEYEHERDMHSAYKHADG------ 166
Query 63 IKVVSKRKNIPGMSRGRG-GGW 83
K + R+ + + RGR GW
Sbjct 167 -KKIDGRRVLVDVERGRTVKGW 187
> mmu:74326 Hnrnpr, 2610003J05Rik, Hnrpr; heterogeneous nuclear
ribonucleoprotein R; K13161 heterogeneous nuclear ribonucleoprotein
R
Length=632
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 24/56 (42%), Positives = 32/56 (57%), Gaps = 5/56 (8%)
Query 6 PADLQEH-----FKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDS 56
P DL E F+ GPI + +M+D +G +GYA+I F + AAQ AV L DS
Sbjct 173 PRDLYEDELVPLFEKAGPIWDLRLMMDPLSGQNRGYAFITFCGKEAAQEAVKLCDS 228
> cel:EEED8.7 rsp-4; SR Protein (splicing factor) family member
(rsp-4); K12891 splicing factor, arginine/serine-rich 2
Length=196
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 39/74 (52%), Gaps = 1/74 (1%)
Query 1 STSATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKD 60
S TP DL+ F+ G I + I DK++ KG+ ++ F A+HA+ +D L D
Sbjct 27 SYQTTPNDLRRTFERYGDIGDVHIPRDKYSRQSKGFGFVRFYERRDAEHALDRTDGKLVD 86
Query 61 -RQIKVVSKRKNIP 73
R+++V + + P
Sbjct 87 GRELRVTLAKYDRP 100
> mmu:53890 Sart3, AU045857, mKIAA0156; squamous cell carcinoma
antigen recognized by T-cells 3
Length=962
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 32/108 (29%), Positives = 50/108 (46%), Gaps = 8/108 (7%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDS-TLKDR 61
S T +L++ K G + + ++ ++ G PKG AY+E+ +E A AV+ D T+++
Sbjct 811 SCTKEELEDICKAHGTVKDLRLVTNR-AGKPKGLAYVEYENESQASQAVMKMDGMTIREN 869
Query 62 QIKVV------SKRKNIPGMSRGRGGGWRPRGAVPFRGRGRGGGSFRP 103
IKV K P + G PR RG+GR S P
Sbjct 870 VIKVAISNPPQRKVPEKPEVRTAPGAPMLPRQMYGARGKGRTQLSLLP 917
Score = 34.3 bits (77), Expect = 0.098, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 35/66 (53%), Gaps = 4/66 (6%)
Query 9 LQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKDRQIKV--- 65
L+ F+ CG + +I + G +GY Y+EFG E +AQ A+ L ++ R + V
Sbjct 722 LRPLFEVCGEVVQIRPIFSN-RGDFRGYCYVEFGEEKSAQQALELDRKIVEGRPMFVSPC 780
Query 66 VSKRKN 71
V K KN
Sbjct 781 VDKSKN 786
> bbo:BBOV_III006220 17.m07552; RNA recognition motif domaining
containing protein; K12831 splicing factor 3B subunit 4
Length=280
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 9 LQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAV 51
L E F GP+ + I DK TGH +GYA++EF ++ A +A+
Sbjct 34 LWEFFVQVGPVKHLHIPRDKVTGHHQGYAFVEFDTDDDADYAI 76
> ath:AT5G44200 CBP20; CBP20 (CAP-BINDING PROTEIN 20); RNA binding
/ RNA cap binding; K12883 nuclear cap-binding protein subunit
2
Length=257
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 46/107 (42%), Gaps = 16/107 (14%)
Query 1 STSATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAV-LLSDSTLK 59
S T L E F G I +I + +DK T P G+ ++ F S + AV +S + L
Sbjct 42 SFYTTEEQLYELFSRAGEIKKIIMGLDKNTKTPCGFCFVLFYSREDTEDAVKYISGTILD 101
Query 60 DRQIKVVSKRKNIPGMSRGRGGGWRPRGAVPFRGRGRGGGSFRPTYR 106
DR I+V G GR W GRGR GG R YR
Sbjct 102 DRPIRVDFD----WGFQEGRQ--W---------GRGRSGGQVRDEYR 133
> tgo:TGME49_070640 RNA binding motif-containing protein
Length=208
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 42/79 (53%), Gaps = 4/79 (5%)
Query 3 SATPADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDSTLKD-R 61
+P +++ F+ G + + + +D FT P+G+ ++EF E AQ A+ D T+ D
Sbjct 15 ETSPDRVRQIFEKFGRVRDVYLPLDHFTKRPRGFGFVEFYEESTAQEAMREMDRTMIDGN 74
Query 62 QIKVV---SKRKNIPGMSR 77
++ V+ +RK+ M R
Sbjct 75 EVHVIIAQDRRKSPETMRR 93
> hsa:10236 HNRNPR, FLJ25714, HNRPR, hnRNP_R, hnRNP-R; heterogeneous
nuclear ribonucleoprotein R; K13161 heterogeneous nuclear
ribonucleoprotein R
Length=532
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 24/56 (42%), Positives = 32/56 (57%), Gaps = 5/56 (8%)
Query 6 PADLQEH-----FKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVLLSDS 56
P DL E F+ GPI + +M+D +G +GYA+I F + AAQ AV L DS
Sbjct 72 PRDLYEDELVPLFEKAGPIWDLRLMMDPLSGQNRGYAFITFCGKEAAQEAVKLCDS 127
> ath:AT3G55460 SCL30; SCL30; RNA binding / nucleic acid binding
/ nucleotide binding
Length=262
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 6 PADLQEHFKGCGPINRITIMVDKFTGHPKGYAYIEFGSEIAAQHAVL-LSDSTLKDRQIK 64
P +L+E F+ GP+ + I D ++G P+G+A++EF A A ++ + R+I
Sbjct 60 PEELREPFERFGPVRDVYIPRDYYSGQPRGFAFVEFVDAYDAGEAQRSMNRRSFAGREIT 119
Query 65 VV 66
VV
Sbjct 120 VV 121
Lambda K H
0.320 0.140 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003209916
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40