bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6959_orf1
Length=186
Score E
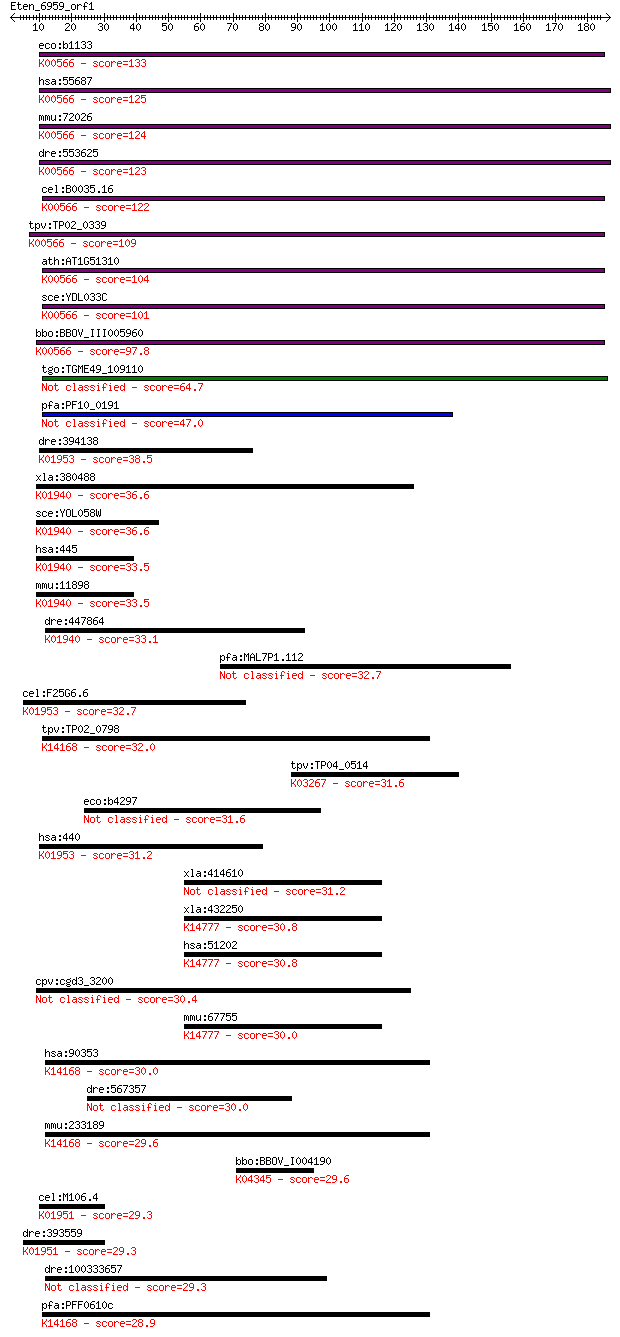
Sequences producing significant alignments: (Bits) Value
eco:b1133 mnmA, asuE, ECK1119, JW1119, trmU, ycfB; tRNA (5-met... 133 3e-31
hsa:55687 TRMU, MGC99627, MTO2, MTU1, TRMT, TRMT1, TRNT1; tRNA... 125 6e-29
mmu:72026 Trmu, 1110005N20Rik, 1600025P05Rik, AI314320, Trmt1;... 124 2e-28
dre:553625 trmu, MGC110555, zgc:110555; tRNA 5-methylaminometh... 123 4e-28
cel:B0035.16 hypothetical protein; K00566 tRNA-specific 2-thio... 122 8e-28
tpv:TP02_0339 tRNA (5-methylaminomethyl-2-thiouridylate)-methy... 109 4e-24
ath:AT1G51310 tRNA (5-methylaminomethyl-2-thiouridylate)-methy... 104 2e-22
sce:YDL033C SLM3, MTO2, MTU1; Slm3p (EC:2.1.1.61); K00566 tRNA... 101 2e-21
bbo:BBOV_III005960 17.m07526; tRNA methyl transferase family p... 97.8 2e-20
tgo:TGME49_109110 tRNA methyltransferase domain-containing pro... 64.7 2e-10
pfa:PF10_0191 tRNA methyltransferase, putative (EC:2.1.1.61) 47.0 3e-05
dre:394138 asns, MGC56605, zgc:56605, zgc:76901; asparagine sy... 38.5 0.012
xla:380488 ass1, MGC53519, ass; argininosuccinate synthase 1 (... 36.6 0.045
sce:YOL058W ARG1, ARG10; Arg1p (EC:6.3.4.5); K01940 argininosu... 36.6 0.049
hsa:445 ASS1, ASS, CTLN1; argininosuccinate synthase 1 (EC:6.3... 33.5 0.45
mmu:11898 Ass1, AA408052, ASS, Ass-1, MGC103151, fold; arginin... 33.5 0.46
dre:447864 ass1, ass, wu:fb95a04, wu:fc01e08, zgc:92051; argin... 33.1 0.52
pfa:MAL7P1.112 conserved Plasmodium protein, unknown function 32.7 0.69
cel:F25G6.6 nrs-2; asparagiNyl tRNA Synthetase family member (... 32.7 0.71
tpv:TP02_0798 hypothetical protein; K14168 cytoplasmic tRNA 2-... 32.0 1.2
tpv:TP04_0514 translation elongation factor EF-1 subunit alpha... 31.6 1.6
eco:b4297 yjhG, ECK4286, JW4259; KpLE2 phage-like element; pre... 31.6 1.7
hsa:440 ASNS, TS11; asparagine synthetase (glutamine-hydrolyzi... 31.2 1.9
xla:414610 hypothetical protein MGC81500 31.2 1.9
xla:432250 ddx47, MGC81303; DEAD (Asp-Glu-Ala-Asp) box polypep... 30.8 2.7
hsa:51202 DDX47, DKFZp564O176, E4-DBP, FLJ30012, HQ0256, MSTP1... 30.8 2.7
cpv:cgd3_3200 hypothetical protein 30.4 3.7
mmu:67755 Ddx47, 4930588A18Rik, C77285; DEAD (Asp-Glu-Ala-Asp)... 30.0 4.7
hsa:90353 CTU1, ATPBD3, MGC17332, NCS6; cytosolic thiouridylas... 30.0 5.0
dre:567357 Sushi, von Willebrand factor type A, EGF and pentra... 30.0 5.2
mmu:233189 Ctu1, Atpbd3, BC005752, MGC6894; cytosolic thiourid... 29.6 6.4
bbo:BBOV_I004190 19.m02221; cAMP-dependent protein kinase (EC:... 29.6 6.9
cel:M106.4 hypothetical protein; K01951 GMP synthase (glutamin... 29.3 7.5
dre:393559 gmps, MGC66002, sb:cb632, wu:fb76b01, wu:fi05a09, z... 29.3 8.0
dre:100333657 plxnb2b; plexin b2b 29.3 9.0
pfa:PFF0610c PP-loop family protein, putative; K14168 cytoplas... 28.9 9.4
> eco:b1133 mnmA, asuE, ECK1119, JW1119, trmU, ycfB; tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase
(EC:2.1.1.61);
K00566 tRNA-specific 2-thiouridylase [EC:2.8.1.-]
Length=368
Score = 133 bits (335), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 78/179 (43%), Positives = 109/179 (60%), Gaps = 6/179 (3%)
Query 10 KKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGGSTAVSDAKK---VADAL 66
KKV V MSGGVDSS++A LL +QGY V G+ M +E E TA +D V D L
Sbjct 6 KKVIVGMSGGVDSSVSAWLLQQQGYQVEGLFMKNWEEDDGEEYCTAAADLADAQAVCDKL 65
Query 67 HIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLE-LGADCVA 125
I + VN+ ++ D+V + F++EY GRTPNP + CN +KF + L+ E LGAD +A
Sbjct 66 GIELHTVNFAAEYWDNVFELFLAEYKAGRTPNPDILCNKEIKFKAFLEFAAEDLGADYIA 125
Query 126 TGHYARIEQDEKTGRYLLKKGLDVRKDQSYVLYTLTQDVLKHFMLPLGNYSKEKTRELA 184
TGHY R + + G+ L +GLD KDQSY LYTL+ + + + P+G K + R++A
Sbjct 126 TGHYVR--RADVDGKSRLLRGLDSNKDQSYFLYTLSHEQIAQSLFPVGELEKPQVRKIA 182
> hsa:55687 TRMU, MGC99627, MTO2, MTU1, TRMT, TRMT1, TRNT1; tRNA
5-methylaminomethyl-2-thiouridylate methyltransferase (EC:2.1.1.61);
K00566 tRNA-specific 2-thiouridylase [EC:2.8.1.-]
Length=421
Score = 125 bits (315), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 77/202 (38%), Positives = 110/202 (54%), Gaps = 25/202 (12%)
Query 10 KKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGGSTA---VSDAKKVADAL 66
+ V A+SGGVDS++ A LL +GY V GV M D E G TA DA +V L
Sbjct 5 RHVVCALSGGVDSAVAALLLRRRGYQVTGVFMKNWDSLDEHGVCTADKDCEDAYRVCQIL 64
Query 67 HIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLE-LGADCVA 125
IP + V+Y K++ +DV F++EY KGRTPNP + CN +KF ++ LGAD +A
Sbjct 65 DIPFHQVSYVKEYWNDVFSDFLNEYEKGRTPNPDIVCNKHIKFSCFFHYAVDNLGADAIA 124
Query 126 TGHYAR--IEQDE-------KTGRYLLKKGLDVR------------KDQSYVLYTLTQDV 164
TGHYAR +E +E K L + +VR KDQ++ L ++QD
Sbjct 125 TGHYARTSLEDEEVFEQKHVKKPEGLFRNRFEVRNAVKLLQAADSFKDQTFFLSQVSQDA 184
Query 165 LKHFMLPLGNYSKEKTRELAGK 186
L+ + PLG +KE +++A +
Sbjct 185 LRRTIFPLGGLTKEFVKKIAAE 206
> mmu:72026 Trmu, 1110005N20Rik, 1600025P05Rik, AI314320, Trmt1;
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase
(EC:2.1.1.61 2.8.1.-); K00566 tRNA-specific 2-thiouridylase
[EC:2.8.1.-]
Length=417
Score = 124 bits (310), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 77/202 (38%), Positives = 110/202 (54%), Gaps = 25/202 (12%)
Query 10 KKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGGSTA---VSDAKKVADAL 66
+ V A+SGGVDS++ A LL +GY V GV M D E+G A DA KV L
Sbjct 5 RHVVCALSGGVDSAVAALLLRRRGYQVTGVFMKNWDSLDEQGVCAADKDCEDAYKVCQIL 64
Query 67 HIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLE-LGADCVA 125
IP + V+Y K++ +DV F++EY KGRTPNP + CN +KF ++ LGAD VA
Sbjct 65 DIPFHQVSYVKEYWNDVFSDFLNEYEKGRTPNPDINCNKHIKFSCFYHYAVDNLGADAVA 124
Query 126 TGHYAR--IEQDE-------KTGRYLLKKGLDVR------------KDQSYVLYTLTQDV 164
TGHYAR +E +E K L + +VR KDQ++ L ++QD
Sbjct 125 TGHYARTSLEDEEVFEQKHTKKPDGLFRNRFEVRNPVKLLQAADSFKDQTFFLSQVSQDA 184
Query 165 LKHFMLPLGNYSKEKTRELAGK 186
L+ + PLG +K+ +++A +
Sbjct 185 LRRTIFPLGELTKDFVKKIAAE 206
> dre:553625 trmu, MGC110555, zgc:110555; tRNA 5-methylaminomethyl-2-thiouridylate
methyltransferase (EC:2.1.1.61 2.8.1.-);
K00566 tRNA-specific 2-thiouridylase [EC:2.8.1.-]
Length=416
Score = 123 bits (308), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 73/202 (36%), Positives = 109/202 (53%), Gaps = 25/202 (12%)
Query 10 KKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGGSTA---VSDAKKVADAL 66
+ V AMSGGVDSS++A LL GY V GV M D E+G ++ DA KV L
Sbjct 5 RHVVCAMSGGVDSSVSALLLKRMGYHVTGVFMKNWDSQEEKGLCSSDRDCEDAYKVCKML 64
Query 67 HIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLE-LGADCVA 125
IP + V+Y K++ +V + EY +GRTPNP + CN +KF + + LGAD +A
Sbjct 65 DIPFHEVSYVKEYWHEVFSNLLWEYERGRTPNPDIICNKHIKFKHFYQYAVNTLGADAMA 124
Query 126 TGHYARIEQDEKT---------------GRYLLKK------GLDVRKDQSYVLYTLTQDV 164
TGHYAR Q+++ R+ ++K G D+ KDQ++ L ++QD
Sbjct 125 TGHYARTSQEDEEVFQQKLTEAPKSLFRDRFEIRKPVRLYQGADLLKDQTFFLSQISQDA 184
Query 165 LKHFMLPLGNYSKEKTRELAGK 186
L+H + PL +K +++A +
Sbjct 185 LRHTLFPLAGLTKGYVKKIAAE 206
> cel:B0035.16 hypothetical protein; K00566 tRNA-specific 2-thiouridylase
[EC:2.8.1.-]
Length=375
Score = 122 bits (306), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 73/186 (39%), Positives = 109/186 (58%), Gaps = 13/186 (6%)
Query 11 KVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGGSTAV-----SDAKKVADA 65
+V + MSGGVDS+++A LL ++G+DVIG+ M ++ + +EEG S SDA+ V D
Sbjct 3 RVVIGMSGGVDSAVSAFLLKKRGFDVIGLHM-INWDVQEEGTSHCPRSKDESDARNVCDR 61
Query 66 LHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLE-LGADCV 124
L+IP + VN+ K++ +DV F+ Y GRT P + CN ++KF K E AD +
Sbjct 62 LNIPFHTVNFVKEYWNDVFLKFLENYKNGRTTVPDIDCNQSIKFDVFHKIAREKFNADFI 121
Query 125 ATGHYAR-----IEQDEK-TGRYLLKKGLDVRKDQSYVLYTLTQDVLKHFMLPLGNYSKE 178
ATGHYA +Q+ K + L G D KDQ++ L T+ Q+ LK M PLG+ K
Sbjct 122 ATGHYATTNFGDFQQNAKDSDEIRLFSGKDPLKDQTFFLCTVNQEQLKRAMFPLGSLQKS 181
Query 179 KTRELA 184
+ + +A
Sbjct 182 EVKRIA 187
> tpv:TP02_0339 tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase;
K00566 tRNA-specific 2-thiouridylase [EC:2.8.1.-]
Length=693
Score = 109 bits (273), Expect = 4e-24, Method: Composition-based stats.
Identities = 67/200 (33%), Positives = 106/200 (53%), Gaps = 22/200 (11%)
Query 7 KMKKKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEG--GSTAVSDAKKVAD 64
+ KK+VAV +SGGVDSSL L+ + +DV + ++D S G S +S A +V +
Sbjct 294 RSKKRVAVLVSGGVDSSLALWLMKSRAFDVHAFYLKVNDLSNGPGLCASNDISYAMQVCN 353
Query 65 ALHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLELGADCV 124
L + +V+ + K + + ++ + +Y KG PNP V CN+ +KFG L+ ++ G D V
Sbjct 354 ILGVTLHVLPFSKAYSEHILSGVLDDYRKGEVPNPDVLCNSRIKFGEFLRMAIDWGFDYV 413
Query 125 ATGHYARIEQDEKT-----------------GRYLLKK---GLDVRKDQSYVLYTLTQDV 164
A+GHYA + D T G Y +K+ D KDQ+Y L LTQ+
Sbjct 414 ASGHYATLCDDFFTKEQIKSPSSLNGSTSINGYYKIKRLCLSNDTLKDQTYFLSRLTQNQ 473
Query 165 LKHFMLPLGNYSKEKTRELA 184
+ + P+G+ +K + RE A
Sbjct 474 MSKLIFPIGHLTKSQVREFA 493
> ath:AT1G51310 tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase;
K00566 tRNA-specific 2-thiouridylase [EC:2.8.1.-]
Length=497
Score = 104 bits (259), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 63/183 (34%), Positives = 97/183 (53%), Gaps = 9/183 (4%)
Query 11 KVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLS-DESREEGGSTAVSD-----AKKVAD 64
+VAV +SGGVDSS+ LL G+ + + E E + + AK V +
Sbjct 83 RVAVLLSGGVDSSVALRLLHAAGHSCTAFYLKIWFQEGFENFWNQCPWEDDLKYAKHVCE 142
Query 65 ALHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLELGADCV 124
+ +P VV+ ++ + V+ Y I EY GRTPNP V CN +KFG+ + ++ D V
Sbjct 143 QVDVPLEVVHLTDEYWERVVSYIIEEYRCGRTPNPDVLCNTRIKFGAFMDAISDMEYDYV 202
Query 125 ATGHYARI---EQDEKTGRYLLKKGLDVRKDQSYVLYTLTQDVLKHFMLPLGNYSKEKTR 181
+GHYA++ D+ +L+ D+ KDQ+Y L L+Q LK + PLG K++ R
Sbjct 203 GSGHYAKVVHPPADQNDASSVLELSQDMVKDQTYFLSHLSQTQLKRLLFPLGCVKKDEVR 262
Query 182 ELA 184
+LA
Sbjct 263 KLA 265
> sce:YDL033C SLM3, MTO2, MTU1; Slm3p (EC:2.1.1.61); K00566 tRNA-specific
2-thiouridylase [EC:2.8.1.-]
Length=417
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 67/185 (36%), Positives = 96/185 (51%), Gaps = 11/185 (5%)
Query 11 KVAVAMSGGVDSSLTAALLIEQGYDVIGVTM-------TLSDESREEGGSTAVSDAKKVA 63
V VAMS GVDSS+ AAL + + GV M +L D +E D +VA
Sbjct 28 NVIVAMSSGVDSSVAAALFAGEFPNTRGVYMQNWSESQSLDDPGKEPCYERDWRDVNRVA 87
Query 64 DALHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSL---LKDCLELG 120
L+I VN+ + + DV + + Y++G TPNP + CN VKFG L L + G
Sbjct 88 KHLNIRVDKVNFEQDYWIDVFEPMLRGYSEGSTPNPDIGCNKFVKFGKLREWLDEKYGTG 147
Query 121 ADCVATGHYARIEQD-EKTGRYLLKKGLDVRKDQSYVLYTLTQDVLKHFMLPLGNYSKEK 179
+ TGHYAR+ Q+ G + L + + KDQSY L + VL +LP+G+ +K +
Sbjct 148 NYWLVTGHYARVMQEMNGKGLFHLLRSIYRPKDQSYYLSQINSTVLSSLLLPIGHLTKPE 207
Query 180 TRELA 184
R+LA
Sbjct 208 VRDLA 212
> bbo:BBOV_III005960 17.m07526; tRNA methyl transferase family
protein; K00566 tRNA-specific 2-thiouridylase [EC:2.8.1.-]
Length=665
Score = 97.8 bits (242), Expect = 2e-20, Method: Composition-based stats.
Identities = 62/184 (33%), Positives = 97/184 (52%), Gaps = 8/184 (4%)
Query 9 KKKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTL-SDESREEGG---STAVSDAKKVAD 64
+ +VA +SGGVDS++ LL +G+D+ + + S E+ + G S + A V
Sbjct 286 QSRVACLVSGGVDSAVALWLLKSRGFDIEAFYLKVWSVENGDLGDCQWSADLEHAMGVTK 345
Query 65 ALHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLELGADCV 124
L++P +VV + + V+ F++ + G TPNP V CN +KFG+ L + G +
Sbjct 346 LLNVPLHVVPFQDVYHQRVLDPFVAGASLGETPNPDVCCNEFIKFGAFLDYAVRWGFSHI 405
Query 125 ATGHYARIEQDEKTGRYL----LKKGLDVRKDQSYVLYTLTQDVLKHFMLPLGNYSKEKT 180
A+GHYA I +D G L+ D+RKDQ+Y L L+Q L+ M PL K +
Sbjct 406 ASGHYASIIEDLVPGTSTRIARLRLCRDLRKDQTYFLSRLSQSQLRRVMFPLSQLYKNEV 465
Query 181 RELA 184
R +A
Sbjct 466 RTIA 469
> tgo:TGME49_109110 tRNA methyltransferase domain-containing protein
(EC:2.1.1.61)
Length=1598
Score = 64.7 bits (156), Expect = 2e-10, Method: Composition-based stats.
Identities = 68/260 (26%), Positives = 102/260 (39%), Gaps = 86/260 (33%)
Query 11 KVAVAMSGGVDSSLTAALLIEQGY------------DVIGVTMTLSD------ESREEGG 52
+VAV +SGGVDSS++ LL ++G+ +++ V+ L+ GG
Sbjct 832 QVAVLLSGGVDSSVSLCLLQQRGFAPQAFFIKVWLPELLLVSRHLNRLLDSGLAPAAAGG 891
Query 53 STAVSD---AKKVADALHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKF 109
D A +V +P V+ + + + V++ + E +G TPNP CN VKF
Sbjct 892 CGWERDLLFADQVCRQARVPLEVLPLQEAYWEGVVQQMLDEARQGLTPNPDWWCNQRVKF 951
Query 110 GSLLKDCLE---------------------LGAD------------CVATGHYARI---- 132
G+ L D L+ AD VA+GHYAR+
Sbjct 952 GAFL-DLLDGRETRFSARRIAGESEGENEKEEADMPFLRNSSRWTGAVASGHYARVVPAA 1010
Query 133 ---------------------------EQDEKTGRYLLKKGLDVRKDQSYVLYTLTQDVL 165
E+ R L +G D RKDQSY L L+Q L
Sbjct 1011 ETSRRSEEGEDTEDRDEDGEEDRGDKERGSEEERRTRLFRGKDRRKDQSYFLSGLSQRQL 1070
Query 166 KHFMLPLGNYSKEKTRELAG 185
+ + P+G+ K + R LA
Sbjct 1071 RRLVTPVGDMEKVEVRRLAA 1090
> pfa:PF10_0191 tRNA methyltransferase, putative (EC:2.1.1.61)
Length=1084
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 36/143 (25%), Positives = 65/143 (45%), Gaps = 21/143 (14%)
Query 11 KVAVAMSGGVDSSLTAALLIEQGY--DVIGVTMTLSDESREEGGSTAVSDAKKVADALHI 68
++A +SGGVDS + LL + + D T +D S+ + + K +
Sbjct 449 RIAHMLSGGVDSLMALHLLERKKFYVDNYFFNFTNADCSKND-----IKYVKDICKNNKR 503
Query 69 PHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLEL--------- 119
+++N + ++ D+V+ + YA G+ PNP + CN +K+ LK +
Sbjct 504 NLFIININDEYFDEVLVPMLFFYADGKVPNPDIMCNQKIKYNFFLKVIKSIYKQKWNYRT 563
Query 120 -----GADCVATGHYARIEQDEK 137
D ++TGHYA I ++K
Sbjct 564 KSKLCNYDFISTGHYAMIRTNDK 586
> dre:394138 asns, MGC56605, zgc:56605, zgc:76901; asparagine
synthetase (EC:6.3.5.4); K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=560
Score = 38.5 bits (88), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 25/70 (35%), Positives = 39/70 (55%), Gaps = 9/70 (12%)
Query 10 KKVAVAMSGGVDSSLTAALLI----EQGYDVIGVTMTLSDESREEGGSTAVSDAKKVADA 65
+++ +SGG+DSSL AALL+ E+ T ++ E S V+ A+KVAD
Sbjct 250 RRIGCLLSGGLDSSLVAALLVKLAKEEKLPYPIQTFSIGAED-----SPDVAAARKVADY 304
Query 66 LHIPHYVVNY 75
+ H+VVN+
Sbjct 305 IGSEHHVVNF 314
> xla:380488 ass1, MGC53519, ass; argininosuccinate synthase 1
(EC:6.3.4.5); K01940 argininosuccinate synthase [EC:6.3.4.5]
Length=411
Score = 36.6 bits (83), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 51/119 (42%), Gaps = 25/119 (21%)
Query 9 KKKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTL-SDESREEGGSTAVS-DAKKVADAL 66
K V +A SGG+D+S L EQG+DVI + +E EE AV+ AKKV
Sbjct 5 KGTVVLAYSGGLDTSCILVWLKEQGFDVIAYLANIGQNEDFEEARKKAVNLGAKKV---- 60
Query 67 HIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLELGADCVA 125
+ +D+ + F+ EY P V+ N + LL L C+A
Sbjct 61 ------------YIEDIRQQFVEEYIW-----PAVQANAIYEDRYLLGTS--LARPCIA 100
> sce:YOL058W ARG1, ARG10; Arg1p (EC:6.3.4.5); K01940 argininosuccinate
synthase [EC:6.3.4.5]
Length=420
Score = 36.6 bits (83), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 9 KKKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDE 46
K KV +A SGG+D+S+ A L++QGY+V+ + E
Sbjct 3 KGKVCLAYSGGLDTSVILAWLLDQGYEVVAFMANVGQE 40
> hsa:445 ASS1, ASS, CTLN1; argininosuccinate synthase 1 (EC:6.3.4.5);
K01940 argininosuccinate synthase [EC:6.3.4.5]
Length=412
Score = 33.5 bits (75), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 16/30 (53%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 9 KKKVAVAMSGGVDSSLTAALLIEQGYDVIG 38
K V +A SGG+D+S L EQGYDVI
Sbjct 4 KGSVVLAYSGGLDTSCILVWLKEQGYDVIA 33
> mmu:11898 Ass1, AA408052, ASS, Ass-1, MGC103151, fold; argininosuccinate
synthetase 1 (EC:6.3.4.5); K01940 argininosuccinate
synthase [EC:6.3.4.5]
Length=412
Score = 33.5 bits (75), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 16/30 (53%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 9 KKKVAVAMSGGVDSSLTAALLIEQGYDVIG 38
K V +A SGG+D+S L EQGYDVI
Sbjct 4 KGSVVLAYSGGLDTSCILVWLKEQGYDVIA 33
> dre:447864 ass1, ass, wu:fb95a04, wu:fc01e08, zgc:92051; argininosuccinate
synthetase 1 (EC:6.3.4.5); K01940 argininosuccinate
synthase [EC:6.3.4.5]
Length=414
Score = 33.1 bits (74), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 14/80 (17%)
Query 12 VAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGGSTAVSDAKKVADALHIPHY 71
V +A SGG+D+S L EQGYDVI L++ ++E A A+K+
Sbjct 6 VVLAYSGGLDTSCILVWLKEQGYDVIAY---LANIGQDEDFDVARKKAEKLGAK------ 56
Query 72 VVNYHKKFKDDVIKYFISEY 91
K F +D+ F+ E+
Sbjct 57 -----KVFIEDLRSEFVQEF 71
> pfa:MAL7P1.112 conserved Plasmodium protein, unknown function
Length=952
Score = 32.7 bits (73), Expect = 0.69, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 45/90 (50%), Gaps = 7/90 (7%)
Query 66 LHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLELGADCVA 125
LH Y++ YH + ++IKY Y+ + C++ NNT SLL D +E G
Sbjct 338 LHCFLYLI-YHYNYFQNIIKYN-DNYSNFYSS--CIKNNNTPFVFSLLLDNIEKGNTDCQ 393
Query 126 TGHYARIEQDEKTGRYLLKKGLDVRKDQSY 155
T +Y + Q+E +K+ L +++Q Y
Sbjct 394 TNYYKELNQNESKN---IKEMLSNQQNQRY 420
> cel:F25G6.6 nrs-2; asparagiNyl tRNA Synthetase family member
(nrs-2); K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=551
Score = 32.7 bits (73), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 39/74 (52%), Gaps = 14/74 (18%)
Query 5 EKKM--KKKVAVAMSGGVDSSLTAAL---LIEQGYDVIGVTMTLSDESREEGGSTAVSDA 59
EK++ + +SGG+DSSL A++ ++Q I ++ D S + +A
Sbjct 222 EKRLMGNRNFGFMLSGGLDSSLIASIATRFLKQ--KPIAFSVGFED-------SPDLENA 272
Query 60 KKVADALHIPHYVV 73
KKVAD L IPH V+
Sbjct 273 KKVADYLKIPHEVL 286
> tpv:TP02_0798 hypothetical protein; K14168 cytoplasmic tRNA
2-thiolation protein 1 [EC:2.7.7.-]
Length=376
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 39/134 (29%), Positives = 62/134 (46%), Gaps = 30/134 (22%)
Query 11 KVAVAMSGGVDSSLTAALL--IEQGYDVIGVTMTLSDESREEGGSTAVSDAKKVADALH- 67
+V V +SGG DSS+ A +L I++ Y G+ L +EG D+ KV ++H
Sbjct 51 RVCVGVSGGKDSSVLAHVLSKIKETY---GLDWNLYLLGVDEGIKGYRDDSLKVVKSMHD 107
Query 68 ---IPHYVVNYHKKF---KDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLELG- 120
I ++++ ++F D V+K G+ N C C GS + LE+G
Sbjct 108 KYKIELKILSFKERFGFSMDQVVKLI------GKKGN-CTVC------GSFRRQMLEIGA 154
Query 121 ----ADCVATGHYA 130
A+ + TGH A
Sbjct 155 RIFEANVLCTGHNA 168
> tpv:TP04_0514 translation elongation factor EF-1 subunit alpha;
K03267 peptide chain release factor subunit 3
Length=539
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 3/55 (5%)
Query 88 ISEYAKGRTPNPCVRCNNTVKFGSLL---KDCLELGADCVATGHYARIEQDEKTG 139
+ + K + PNP NN++ LL CLE ADC G + ++D+ G
Sbjct 474 VDKATKKKKPNPVFVSNNSIVTAHLLVTPAACLEKFADCPQLGRFTLRDEDKTIG 528
> eco:b4297 yjhG, ECK4286, JW4259; KpLE2 phage-like element; predicted
dehydratase
Length=655
Score = 31.6 bits (70), Expect = 1.7, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 42/94 (44%), Gaps = 21/94 (22%)
Query 24 LTAALLIEQ-GYDVIGVTMTLS-----DESREEG-------GSTAVSDAKKVADALHIPH 70
LTA +LI + D+ G+TM DE +G G +D K VA ALH H
Sbjct 30 LTAEMLINRPSGDLFGMTMNAGMGWSPDELDRDGILLLSTLGGLRGADGKPVALALHQGH 89
Query 71 YVVNYHKKFKDDVIK--------YFISEYAKGRT 96
Y ++ K +VIK ++S+ GRT
Sbjct 90 YELDIQMKAAAEVIKANHALPYAVYVSDPCDGRT 123
> hsa:440 ASNS, TS11; asparagine synthetase (glutamine-hydrolyzing)
(EC:6.3.5.4); K01953 asparagine synthase (glutamine-hydrolysing)
[EC:6.3.5.4]
Length=540
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 10 KKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGGSTAVSDAKKVADALHIP 69
+++ +SGG+DSSL AA L++Q + V L + S + A+KVAD +
Sbjct 230 RRIGCLLSGGLDSSLVAATLLKQLKEA-QVQYPLQTFAIGMEDSPDLLAARKVADHIGSE 288
Query 70 HYVVNYHKK 78
HY V ++ +
Sbjct 289 HYEVLFNSE 297
> xla:414610 hypothetical protein MGC81500
Length=317
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 55 AVSDAKKVADALHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLK 114
AVS + + L + V KFKD + Y ++E A C CNNT + LL+
Sbjct 222 AVSSKYQTVEKLQ--QFYVFIPSKFKDSYLVYILNELAGNSFMIFCSTCNNTQRVALLLR 279
Query 115 D 115
+
Sbjct 280 N 280
> xla:432250 ddx47, MGC81303; DEAD (Asp-Glu-Ala-Asp) box polypeptide
47; K14777 ATP-dependent RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=448
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 55 AVSDAKKVADALHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLK 114
AVS + + L + V KFKD + Y ++E A C CNNT + LL+
Sbjct 222 AVSSKYQTVEKLQ--QFYVFIPSKFKDSYLVYILNELAGNSFMIFCSTCNNTQRVALLLR 279
Query 115 D 115
+
Sbjct 280 N 280
> hsa:51202 DDX47, DKFZp564O176, E4-DBP, FLJ30012, HQ0256, MSTP162,
RRP3; DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (EC:3.6.4.13);
K14777 ATP-dependent RNA helicase DDX47/RRP3 [EC:3.6.4.13]
Length=455
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 55 AVSDAKKVADALHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLK 114
AVS + + L Y + KFKD + Y ++E A C CNNT + LL+
Sbjct 227 AVSSKYQTVEKLQ--QYYIFIPSKFKDTYLVYILNELAGNSFMIFCSTCNNTQRTALLLR 284
Query 115 D 115
+
Sbjct 285 N 285
> cpv:cgd3_3200 hypothetical protein
Length=394
Score = 30.4 bits (67), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 32/125 (25%), Positives = 55/125 (44%), Gaps = 11/125 (8%)
Query 9 KKKVAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGGST----AVSDAKKVAD 64
K+KV + + GGV S A I+Q DV+G S + G ST + + + D
Sbjct 240 KRKVGIKLQGGVSSLDIAREFIKQVEDVLGNEYVSSPNTFRIGSSTLHPLIIQELLGIDD 299
Query 65 ALHIPHYV----VNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNT-VKFGSLLKDCLEL 119
L I H + +N + ++D I +E P+ + N T K GS ++ +++
Sbjct 300 ELRIVHTLGIKPINAEQVNQEDNISSPENELVGISHPSAPISSNQTREKIGS--QEVIDI 357
Query 120 GADCV 124
D +
Sbjct 358 NKDII 362
> mmu:67755 Ddx47, 4930588A18Rik, C77285; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 47 (EC:3.6.4.13); K14777 ATP-dependent RNA
helicase DDX47/RRP3 [EC:3.6.4.13]
Length=455
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 55 AVSDAKKVADALHIPHYVVNYHKKFKDDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLK 114
AVS + + L Y + KFKD + Y ++E A C CNNT + LL+
Sbjct 227 AVSSKYQTVEKLQ--QYYLFIPSKFKDTYLVYILNELAGNSFMIFCSTCNNTQRTALLLR 284
Query 115 D 115
+
Sbjct 285 N 285
> hsa:90353 CTU1, ATPBD3, MGC17332, NCS6; cytosolic thiouridylase
subunit 1 homolog (S. pombe); K14168 cytoplasmic tRNA 2-thiolation
protein 1 [EC:2.7.7.-]
Length=348
Score = 30.0 bits (66), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 54/122 (44%), Gaps = 5/122 (4%)
Query 12 VAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGG--STAVSDAKKVADALHIP 69
VAV SGG DS++ A +L +G+++ L GG A++ ++ A +P
Sbjct 54 VAVGASGGKDSTVLAHVLRALAPR-LGISLQLVAVDEGIGGYRDAALAAVRRQAARWELP 112
Query 70 HYVVNYHKKFKDDVIKYFI-SEYAKGRTPNPCVRCNNTVKFGSLLKDCLELGADCVATGH 128
VV Y F + S GR+ + C C ++ +L + +GA + TGH
Sbjct 113 LTVVAYEDLFGGWTMDAVARSTAGSGRSRSCCTFC-GVLRRRALEEGARRVGATHIVTGH 171
Query 129 YA 130
A
Sbjct 172 NA 173
> dre:567357 Sushi, von Willebrand factor type A, EGF and pentraxin
domain-containing protein 1-like
Length=3644
Score = 30.0 bits (66), Expect = 5.2, Method: Composition-based stats.
Identities = 20/63 (31%), Positives = 27/63 (42%), Gaps = 0/63 (0%)
Query 25 TAALLIEQGYDVIGVTMTLSDESREEGGSTAVSDAKKVADALHIPHYVVNYHKKFKDDVI 84
TA + GYD++G + L +S + G V K A IP+ V Y K I
Sbjct 2499 TAVYSCKAGYDILGNSTVLCGQSGQWIGGIPVCHPVKCAAPKEIPNGSVRYSKLQFSQSI 2558
Query 85 KYF 87
YF
Sbjct 2559 TYF 2561
> mmu:233189 Ctu1, Atpbd3, BC005752, MGC6894; cytosolic thiouridylase
subunit 1 homolog (S. pombe); K14168 cytoplasmic tRNA
2-thiolation protein 1 [EC:2.7.7.-]
Length=420
Score = 29.6 bits (65), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 53/127 (41%), Gaps = 15/127 (11%)
Query 12 VAVAMSGGVDSSLTAALLIEQGYDVIGVTMTLSDESREEGG--STAVSDAKKVADALHIP 69
VAV SGG DS++ A +L E +G+T+ L GG A+ + A +P
Sbjct 54 VAVGASGGKDSTVLAHVLRELAPR-LGITLHLVAVDEGIGGYRDAALEAVRSQAARWELP 112
Query 70 HYVVNYHKKFKDDVIKYFI-SEYAKGRTPNPCVRCNNTVKFGSLLKDCLE-----LGADC 123
+V Y F + S GR+ + C C G L + LE +GA
Sbjct 113 LTIVAYEDLFGGWTMDAVARSTAGSGRSRSCCTFC------GVLRRRALEEGARLVGATH 166
Query 124 VATGHYA 130
+ TGH A
Sbjct 167 IVTGHNA 173
> bbo:BBOV_I004190 19.m02221; cAMP-dependent protein kinase (EC:2.7.11.1);
K04345 protein kinase A [EC:2.7.11.11]
Length=416
Score = 29.6 bits (65), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 71 YVVNYHKKFKDDVIKYFISEYAKG 94
++VNY K F+DD+ Y + EY G
Sbjct 167 FIVNYVKSFQDDINVYILLEYVSG 190
> cel:M106.4 hypothetical protein; K01951 GMP synthase (glutamine-hydrolysing)
[EC:6.3.5.2]
Length=792
Score = 29.3 bits (64), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 14/20 (70%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 10 KKVAVAMSGGVDSSLTAALL 29
KKV V +SGGVDS++ AALL
Sbjct 320 KKVLVMVSGGVDSAVCAALL 339
> dre:393559 gmps, MGC66002, sb:cb632, wu:fb76b01, wu:fi05a09,
zgc:66002; guanine monphosphate synthetase (EC:6.3.5.2); K01951
GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2]
Length=692
Score = 29.3 bits (64), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 15/25 (60%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 5 EKKMKKKVAVAMSGGVDSSLTAALL 29
EK K KV V +SGGVDS++ ALL
Sbjct 231 EKVDKSKVLVLLSGGVDSTVCTALL 255
> dre:100333657 plxnb2b; plexin b2b
Length=1811
Score = 29.3 bits (64), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 20/91 (21%), Positives = 39/91 (42%), Gaps = 4/91 (4%)
Query 12 VAVAMSGGVDSSLTAALLIEQGYDVIG----VTMTLSDESREEGGSTAVSDAKKVADALH 67
V V + +D+ + L+ +GY + ++ + R+ ++ +A V
Sbjct 150 VGVMATLEIDNKPFSVFLVGKGYGSMDSSKLISTRIVQNYRDWVVFESIIEASTVQAVPF 209
Query 68 IPHYVVNYHKKFKDDVIKYFISEYAKGRTPN 98
+P Y+ ++ FK D YF+ GRT N
Sbjct 210 VPKYLHDFRFAFKSDNFVYFLFSRTLGRTDN 240
> pfa:PFF0610c PP-loop family protein, putative; K14168 cytoplasmic
tRNA 2-thiolation protein 1 [EC:2.7.7.-]
Length=482
Score = 28.9 bits (63), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 36/129 (27%), Positives = 58/129 (44%), Gaps = 20/129 (15%)
Query 11 KVAVAMSGGVDSSLTAALLI--EQGYDVIGVTMTLSDESREEGGSTAVSDAKKVADAL-- 66
K+ +A+SGG DSS+ A +L+ ++ Y+ L + +EG D+ KV L
Sbjct 50 KICIAVSGGKDSSVLAHVLVNLKKKYNYNWELFLL---AIDEGIKGYRDDSLKVVYKLEK 106
Query 67 --HIPHYVVNYHKKFK---DDVIKYFISEYAKGRTPNPCVRCNNTVKFGSLLKDCLELGA 121
++P ++ + F DDV+K+ G+ N C C + S K L A
Sbjct 107 LYNLPLKILKFENLFSYTMDDVVKFI------GK-KNNCTVC-GVFRRQSFEKGALLFNA 158
Query 122 DCVATGHYA 130
+ TGH A
Sbjct 159 TKLVTGHNA 167
Lambda K H
0.317 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40