bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6872_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
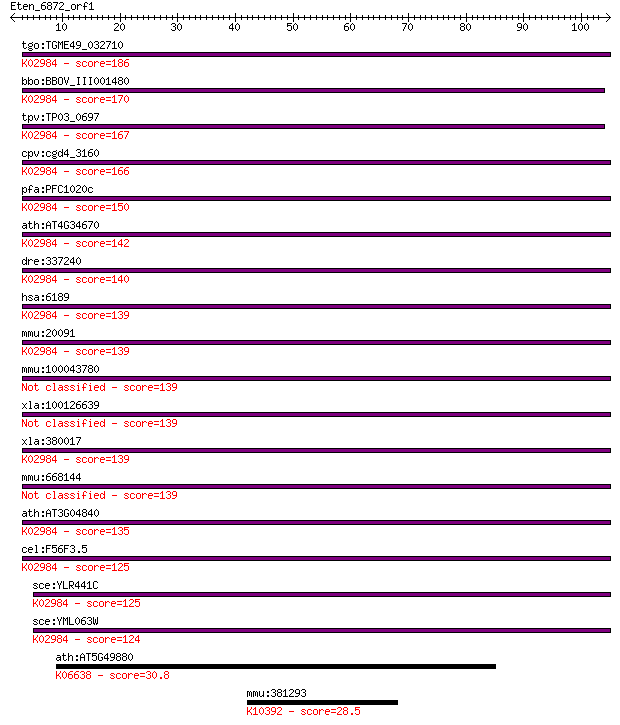
tgo:TGME49_032710 40S ribosomal protein S3a, putative ; K02984... 186 1e-47
bbo:BBOV_III001480 17.m07150; 40S ribosomal protein S3Ae; K029... 170 9e-43
tpv:TP03_0697 40S ribosomal protein S3A; K02984 small subunit ... 167 1e-41
cpv:cgd4_3160 40S ribosomal protein S3A ; K02984 small subunit... 166 2e-41
pfa:PFC1020c 40S ribosomal protein S3A, putative; K02984 small... 150 9e-37
ath:AT4G34670 40S ribosomal protein S3A (RPS3aB); K02984 small... 142 2e-34
dre:337240 rps3a, MGC86672, fb02h01, rpS3Ae, wu:fb02h01, zgc:7... 140 1e-33
hsa:6189 RPS3A, FTE1, MFTL, MGC23240; ribosomal protein S3A; K... 139 2e-33
mmu:20091 Rps3a, MGC102469, MGC117914; ribosomal protein S3A; ... 139 2e-33
mmu:100043780 Gm10119; predicted gene 10119 139 2e-33
xla:100126639 rps3a-b, fte1, mftl; ribosomal protein S3A 139 2e-33
xla:380017 rps3a-a, MGC53968, fte1, mftl, rps3a; ribosomal pro... 139 2e-33
mmu:668144 Gm9000, EG668144; predicted gene 9000 139 2e-33
ath:AT3G04840 40S ribosomal protein S3A (RPS3aA); K02984 small... 135 5e-32
cel:F56F3.5 rps-1; Ribosomal Protein, Small subunit family mem... 125 3e-29
sce:YLR441C RPS1A, RP10A; Rps1ap; K02984 small subunit ribosom... 125 5e-29
sce:YML063W RPS1B, PLC2, RP10B; Rps1bp; K02984 small subunit r... 124 8e-29
ath:AT5G49880 mitotic checkpoint family protein; K06638 mitoti... 30.8 1.2
mmu:381293 Kif14, D1Ertd367e, E130203M01, MGC189899; kinesin f... 28.5 5.6
> tgo:TGME49_032710 40S ribosomal protein S3a, putative ; K02984
small subunit ribosomal protein S3Ae
Length=259
Score = 186 bits (473), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 86/102 (84%), Positives = 96/102 (94%), Gaps = 0/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
+LAVD LRGRVYEVNLADLN DEDQGFRKIKL CE+IQGR+CLTDFHGM++TRDK+CSL+
Sbjct 55 KLAVDGLRGRVYEVNLADLNNDEDQGFRKIKLCCEDIQGRNCLTDFHGMDMTRDKICSLV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
RK HSLIEAFVDV T+DGYTLRMFCIAFT+RRP Q+KSTCYA
Sbjct 115 RKCHSLIEAFVDVTTLDGYTLRMFCIAFTKRRPEQVKSTCYA 156
> bbo:BBOV_III001480 17.m07150; 40S ribosomal protein S3Ae; K02984
small subunit ribosomal protein S3Ae
Length=264
Score = 170 bits (431), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 76/101 (75%), Positives = 90/101 (89%), Gaps = 0/101 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
+LA D LRGR+YEV+LADLN DEDQ +RKIKL CE+IQGR+CLT+FHG ++TRDKLCSLI
Sbjct 55 KLATDGLRGRIYEVSLADLNADEDQSYRKIKLCCEDIQGRNCLTNFHGTSVTRDKLCSLI 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCY 103
RK +SLIEAF DV+T DGYTLRMFCI +T+RRP Q+KSTCY
Sbjct 115 RKNYSLIEAFTDVKTTDGYTLRMFCIGYTKRRPGQLKSTCY 155
> tpv:TP03_0697 40S ribosomal protein S3A; K02984 small subunit
ribosomal protein S3Ae
Length=262
Score = 167 bits (422), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 71/101 (70%), Positives = 90/101 (89%), Gaps = 0/101 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
+LA D L+GRV+E++LADLN DEDQ +RKIKL CE++QGR+CLTDFHG ++TRDKLCSLI
Sbjct 55 KLATDGLKGRVFEMSLADLNADEDQAYRKIKLSCEDVQGRNCLTDFHGTSVTRDKLCSLI 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCY 103
RK +SLIEAF DV+T+DGY LR+FC+ +T+RRP Q+KSTCY
Sbjct 115 RKNYSLIEAFADVKTLDGYNLRLFCVGYTKRRPGQLKSTCY 155
> cpv:cgd4_3160 40S ribosomal protein S3A ; K02984 small subunit
ribosomal protein S3Ae
Length=262
Score = 166 bits (420), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 76/102 (74%), Positives = 91/102 (89%), Gaps = 0/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
+LAVD+LR RVYEV+LADLN DEDQ FRKIKLQCE+IQGR+CLT+FHGM++TRDKLCSLI
Sbjct 55 KLAVDALRHRVYEVSLADLNNDEDQAFRKIKLQCEDIQGRTCLTEFHGMDMTRDKLCSLI 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
RK+ +LIEA +V+T DGY LR+FCIAFT R +Q+KSTCYA
Sbjct 115 RKYQTLIEAHCNVKTSDGYVLRLFCIAFTRRMADQVKSTCYA 156
> pfa:PFC1020c 40S ribosomal protein S3A, putative; K02984 small
subunit ribosomal protein S3Ae
Length=262
Score = 150 bits (379), Expect = 9e-37, Method: Compositional matrix adjust.
Identities = 70/102 (68%), Positives = 83/102 (81%), Gaps = 0/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
+LA DSL+GR+YEVNLADLN DEDQ +KIKL C+ I R C TDF G++ITRDKLCSLI
Sbjct 55 KLATDSLKGRIYEVNLADLNNDEDQAHKKIKLSCDHIINRDCYTDFCGLSITRDKLCSLI 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
RK ++LIE DV+T+D Y LRMFCIAFT++R NQ KSTCYA
Sbjct 115 RKGYTLIEGHTDVKTLDNYHLRMFCIAFTKKRQNQTKSTCYA 156
> ath:AT4G34670 40S ribosomal protein S3A (RPS3aB); K02984 small
subunit ribosomal protein S3Ae
Length=262
Score = 142 bits (359), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 64/102 (62%), Positives = 83/102 (81%), Gaps = 0/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A + L+ RV+EV+LADL DED +RKI+L+ E++QGR+ LT F GM+ T DKL SL+
Sbjct 55 KIASEGLKHRVFEVSLADLQNDEDNAYRKIRLRAEDVQGRNVLTQFWGMDFTTDKLRSLV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KW +LIEA VDV+T DGYTLRMFCIAFT+RR NQ+K TCYA
Sbjct 115 KKWQTLIEAHVDVKTTDGYTLRMFCIAFTKRRANQVKRTCYA 156
> dre:337240 rps3a, MGC86672, fb02h01, rpS3Ae, wu:fb02h01, zgc:73195,
zgc:86672; ribosomal protein S3A; K02984 small subunit
ribosomal protein S3Ae
Length=267
Score = 140 bits (352), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 60/102 (58%), Positives = 84/102 (82%), Gaps = 1/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A D L+GRV+EV+LADL DE FRK KL E++QG++CLT+FHGM++TRDK+CS++
Sbjct 56 RIASDGLKGRVFEVSLADLQNDE-VAFRKFKLVTEDVQGKNCLTNFHGMDLTRDKMCSMV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KW ++IEA VDV+T DGY LR+FC+ FT++R NQI+ T YA
Sbjct 115 KKWQTMIEAHVDVKTTDGYLLRLFCVGFTKKRTNQIRKTSYA 156
> hsa:6189 RPS3A, FTE1, MFTL, MGC23240; ribosomal protein S3A;
K02984 small subunit ribosomal protein S3Ae
Length=264
Score = 139 bits (351), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 60/102 (58%), Positives = 84/102 (82%), Gaps = 1/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A D L+GRV+EV+LADL DE FRK KL E++QG++CLT+FHGM++TRDK+CS++
Sbjct 56 KIASDGLKGRVFEVSLADLQNDE-VAFRKFKLITEDVQGKNCLTNFHGMDLTRDKMCSMV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KW ++IEA VDV+T DGY LR+FC+ FT++R NQI+ T YA
Sbjct 115 KKWQTMIEAHVDVKTTDGYLLRLFCVGFTKKRNNQIRKTSYA 156
> mmu:20091 Rps3a, MGC102469, MGC117914; ribosomal protein S3A;
K02984 small subunit ribosomal protein S3Ae
Length=264
Score = 139 bits (351), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 60/102 (58%), Positives = 84/102 (82%), Gaps = 1/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A D L+GRV+EV+LADL DE FRK KL E++QG++CLT+FHGM++TRDK+CS++
Sbjct 56 KIASDGLKGRVFEVSLADLQNDE-VAFRKFKLITEDVQGKNCLTNFHGMDLTRDKMCSMV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KW ++IEA VDV+T DGY LR+FC+ FT++R NQI+ T YA
Sbjct 115 KKWQTMIEAHVDVKTTDGYLLRLFCVGFTKKRNNQIRKTSYA 156
> mmu:100043780 Gm10119; predicted gene 10119
Length=264
Score = 139 bits (351), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 60/102 (58%), Positives = 84/102 (82%), Gaps = 1/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A D L+GRV+EV+LADL DE FRK KL E++QG++CLT+FHGM++TRDK+CS++
Sbjct 56 KIASDGLKGRVFEVSLADLQNDE-VAFRKFKLITEDVQGKNCLTNFHGMDLTRDKMCSMV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KW ++IEA VDV+T DGY LR+FC+ FT++R NQI+ T YA
Sbjct 115 KKWQTMIEAHVDVKTTDGYLLRLFCVGFTKKRNNQIRKTSYA 156
> xla:100126639 rps3a-b, fte1, mftl; ribosomal protein S3A
Length=264
Score = 139 bits (351), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 60/102 (58%), Positives = 84/102 (82%), Gaps = 1/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A D L+GRV+EV+LADL DE FRK KL E++QG++CLT+FHGM++TRDK+CS++
Sbjct 56 KIASDGLKGRVFEVSLADLQNDE-VAFRKFKLITEDVQGKNCLTNFHGMDLTRDKMCSMV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KW ++IEA VDV+T DGY LR+FC+ FT++R NQI+ T YA
Sbjct 115 KKWQTMIEAHVDVKTTDGYLLRLFCVGFTKKRNNQIRKTSYA 156
> xla:380017 rps3a-a, MGC53968, fte1, mftl, rps3a; ribosomal protein
S3A; K02984 small subunit ribosomal protein S3Ae
Length=264
Score = 139 bits (350), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 60/102 (58%), Positives = 84/102 (82%), Gaps = 1/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A D L+GRV+EV+LADL DE FRK KL E++QG++CLT+FHGM++TRDK+CS++
Sbjct 56 KIASDGLKGRVFEVSLADLQNDE-VAFRKFKLITEDVQGKNCLTNFHGMDLTRDKMCSMV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KW ++IEA VDV+T DGY LR+FC+ FT++R NQI+ T YA
Sbjct 115 KKWQTMIEAHVDVKTTDGYLLRLFCVGFTKKRNNQIRKTSYA 156
> mmu:668144 Gm9000, EG668144; predicted gene 9000
Length=264
Score = 139 bits (350), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 59/102 (57%), Positives = 84/102 (82%), Gaps = 1/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A D L+GRV+EV+LADL DE FRK KL E++QG++CLT+FHGM++TRDK+CS++
Sbjct 56 KIASDGLKGRVFEVSLADLQNDE-VAFRKFKLTTEDVQGKNCLTNFHGMDLTRDKMCSMV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KW ++IEA VDV+T DGY LR+FC+ F+++R NQI+ T YA
Sbjct 115 KKWQTMIEAHVDVKTTDGYLLRLFCVGFSKKRNNQIRKTSYA 156
> ath:AT3G04840 40S ribosomal protein S3A (RPS3aA); K02984 small
subunit ribosomal protein S3Ae
Length=262
Score = 135 bits (339), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 61/102 (59%), Positives = 81/102 (79%), Gaps = 0/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A + L+ RV+EV+LADL DED +RKI+L+ E++QGR+ L F GM+ T DKL SL+
Sbjct 55 KIASEGLKHRVFEVSLADLQGDEDNAYRKIRLRAEDVQGRNVLCQFWGMDFTTDKLRSLV 114
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KW +LIEA VDV+T D YTLR+FCIAFT+RR NQ+K TCYA
Sbjct 115 KKWQTLIEAHVDVKTTDSYTLRLFCIAFTKRRANQVKRTCYA 156
> cel:F56F3.5 rps-1; Ribosomal Protein, Small subunit family member
(rps-1); K02984 small subunit ribosomal protein S3Ae
Length=257
Score = 125 bits (314), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 55/102 (53%), Positives = 79/102 (77%), Gaps = 1/102 (0%)
Query 3 QLAVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLI 62
++A + L+GRV+EV+L DLN + + FRK KL E++QG++ LT+FH M++T DKLCS++
Sbjct 54 KIASEGLKGRVFEVSLGDLN-NSEADFRKFKLIAEDVQGKNVLTNFHAMSMTHDKLCSIV 112
Query 63 RKWHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
+KWH+LIEA V+T DGYTLR+F IAFT++ NQ+K T Y
Sbjct 113 KKWHTLIEANTAVKTTDGYTLRVFVIAFTKKSVNQVKKTSYT 154
> sce:YLR441C RPS1A, RP10A; Rps1ap; K02984 small subunit ribosomal
protein S3Ae
Length=255
Score = 125 bits (313), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 58/100 (58%), Positives = 75/100 (75%), Gaps = 0/100 (0%)
Query 5 AVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLIRK 64
A D+L+GRV EV LADL ED FRKIKL+ +E+QG++ LT+FHGM+ T DKL S++RK
Sbjct 57 ASDALKGRVVEVCLADLQGSEDHSFRKIKLRVDEVQGKNLLTNFHGMDFTTDKLRSMVRK 116
Query 65 WHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
W +LIEA V V+T D Y LR+F IAFT ++ NQ+K YA
Sbjct 117 WQTLIEANVTVKTSDDYVLRIFAIAFTRKQANQVKRHSYA 156
> sce:YML063W RPS1B, PLC2, RP10B; Rps1bp; K02984 small subunit
ribosomal protein S3Ae
Length=255
Score = 124 bits (311), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 57/100 (57%), Positives = 75/100 (75%), Gaps = 0/100 (0%)
Query 5 AVDSLRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLIRK 64
A D+L+GRV EV LADL ED FRK+KL+ +E+QG++ LT+FHGM+ T DKL S++RK
Sbjct 57 ASDALKGRVVEVCLADLQGSEDHSFRKVKLRVDEVQGKNLLTNFHGMDFTTDKLRSMVRK 116
Query 65 WHSLIEAFVDVQTVDGYTLRMFCIAFTERRPNQIKSTCYA 104
W +LIEA V V+T D Y LR+F IAFT ++ NQ+K YA
Sbjct 117 WQTLIEANVTVKTSDDYVLRIFAIAFTRKQANQVKRHSYA 156
> ath:AT5G49880 mitotic checkpoint family protein; K06638 mitotic
spindle assembly checkpoint protein MAD1
Length=726
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 36/76 (47%), Gaps = 8/76 (10%)
Query 9 LRGRVYEVNLADLNLDEDQGFRKIKLQCEEIQGRSCLTDFHGMNITRDKLCSLIRKWHSL 68
L+ R + L ++NL E+Q R+ + S L+ FH + ++ +KL + + W SL
Sbjct 298 LKSRHLDAELLNVNLLEEQSRRE--------RAESELSKFHDLQLSMEKLENELSSWKSL 349
Query 69 IEAFVDVQTVDGYTLR 84
+ V D +R
Sbjct 350 LNDIPGVSCPDDIVMR 365
> mmu:381293 Kif14, D1Ertd367e, E130203M01, MGC189899; kinesin
family member 14; K10392 kinesin family member 1/13/14
Length=1624
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 42 RSCLTDFHGMNITRDKLCSLIRKWHS 67
R C +D H + + +CSL RK HS
Sbjct 1501 RGCPSDLHCLRSCTETICSLARKLHS 1526
Lambda K H
0.327 0.140 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022291452
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40