bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6811_orf2
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
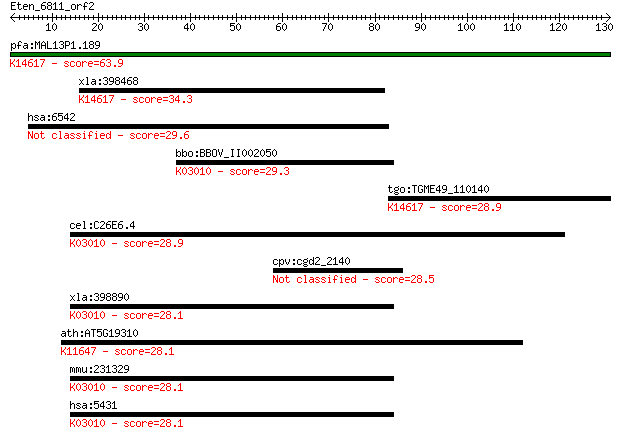
pfa:MAL13P1.189 conserved Plasmodium membrane protein, unknown... 63.9 1e-10
xla:398468 lmbrd1; LMBR1 domain containing 1; K14617 LMBR1 dom... 34.3 0.089
hsa:6542 SLC7A2, ATRC2, CAT2, HCAT2; solute carrier family 7 (... 29.6 2.4
bbo:BBOV_II002050 18.m06162; DNA-directed RNA polymerase, beta... 29.3 3.5
tgo:TGME49_110140 hypothetical protein ; K14617 LMBR1 domain-c... 28.9 3.9
cel:C26E6.4 rpb-2; RNA Polymerase II (B) subunit family member... 28.9 4.6
cpv:cgd2_2140 hypothetical protein 28.5 5.4
xla:398890 polr2b, MGC68569; polymerase (RNA) II (DNA directed... 28.1 6.3
ath:AT5G19310 homeotic gene regulator, putative; K11647 SWI/SN... 28.1 7.3
mmu:231329 Polr2b, MGC47046, Pol2rb, Rpb140, Rpb2; polymerase ... 28.1 7.8
hsa:5431 POLR2B, POL2RB, RPB2, hRPB140, hsRPB2; polymerase (RN... 28.1 7.8
> pfa:MAL13P1.189 conserved Plasmodium membrane protein, unknown
function; K14617 LMBR1 domain-containing protein 1
Length=530
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 33/131 (25%), Positives = 67/131 (51%), Gaps = 1/131 (0%)
Query 1 QQRQLQIKYAGGLH-EMRPEDSEALEKLRVQQRLCSERNYKLQEAEQAHRSAYSLLCRLM 59
+++++ IK ++ E++ D + + K+ +R+ S+ NYKLQE E+ S S L ++
Sbjct 231 KEKEIMIKNKYNINCEIKETDKDEMLKIEKMKRVLSKYNYKLQEIEKLSESWLSYLIGIV 290
Query 60 LPFRKTVGLVLLLLLLLLFVSLGTVLIRRLEGSTCGWSCGFATQSSEGPSTPANAALLFL 119
FR G++ L ++++SL + + S C + CGF + + L+F
Sbjct 291 FTFRLLTGMIFLAFSFIIYISLLASITDKYFNSICAYKCGFVLEQINTIFNLLDTLLIFF 350
Query 120 SSYPPTDLWLV 130
S + P D+ ++
Sbjct 351 SKFFPLDILII 361
> xla:398468 lmbrd1; LMBR1 domain containing 1; K14617 LMBR1 domain-containing
protein 1
Length=537
Score = 34.3 bits (77), Expect = 0.089, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 36/66 (54%), Gaps = 3/66 (4%)
Query 16 MRPEDSEALEKLRVQQRLCSERNYKLQEAEQAHRSAYSLLCRLMLPFRKTVGLVLLLLLL 75
+ +D +AL KL+ + R ++ L+ E + ++ C +M PF+ G++ +L+ L
Sbjct 259 LSSKDRQALYKLQEKLRTLKRKDRHLEHHEN---NCWTKCCLVMRPFKIVWGILFILVAL 315
Query 76 LLFVSL 81
L VSL
Sbjct 316 LFIVSL 321
> hsa:6542 SLC7A2, ATRC2, CAT2, HCAT2; solute carrier family 7
(cationic amino acid transporter, y+ system), member 2; K13864
solute carrier family 7 (cationic amino acid transporter),
member 2
Length=658
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 39/78 (50%), Gaps = 0/78 (0%)
Query 5 LQIKYAGGLHEMRPEDSEALEKLRVQQRLCSERNYKLQEAEQAHRSAYSLLCRLMLPFRK 64
L ++Y GL +P+ S + L R+ S+ ++ ++ S +L C +LP ++
Sbjct 430 LILRYQPGLSYDQPKCSPEKDGLGSSPRVTSKSESQVTMLQRQGFSMRTLFCPSLLPTQQ 489
Query 65 TVGLVLLLLLLLLFVSLG 82
+ LV L+ L F+ LG
Sbjct 490 SASLVSFLVGFLAFLVLG 507
> bbo:BBOV_II002050 18.m06162; DNA-directed RNA polymerase, beta
subunit family protein (EC:2.7.7.6); K03010 DNA-directed
RNA polymerase II subunit RPB2 [EC:2.7.7.6]
Length=1207
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Query 37 RNYKLQEAEQAHRSAYSLLCRLMLPFRKTVGLVLLLLLLLLFVSLGT 83
R K+ + Q H + + ++C P + VGLV L L+ F+S+GT
Sbjct 534 REGKIAKPRQLHNTHWGMICPAETPEGQAVGLV-KNLALMCFISVGT 579
> tgo:TGME49_110140 hypothetical protein ; K14617 LMBR1 domain-containing
protein 1
Length=728
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 26/53 (49%), Gaps = 5/53 (9%)
Query 83 TVLIRRLEGSTCGWSCGF-----ATQSSEGPSTPANAALLFLSSYPPTDLWLV 130
T ++RLE S C +SCGF AT P + L+ LS + P D LV
Sbjct 475 TYRLQRLEFSRCTYSCGFVLDDDATAREGVLFNPLDELLVRLSQFFPADFLLV 527
> cel:C26E6.4 rpb-2; RNA Polymerase II (B) subunit family member
(rpb-2); K03010 DNA-directed RNA polymerase II subunit RPB2
[EC:2.7.7.6]
Length=1193
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 31/118 (26%), Positives = 53/118 (44%), Gaps = 18/118 (15%)
Query 14 HEMRPEDSEALEKLRVQQRLCSERNY--------KLQEAEQAHRSAYSLLCRLMLPFRKT 65
H+ R S+ L +L L R KL + Q H + + ++C P +
Sbjct 466 HQSRAGVSQVLNRLTYTATLSHLRRANSPIGREGKLAKPRQLHNTQWGMVCPAETPEGQA 525
Query 66 VGLVLLLLLLLLFVSLGTV---LIRRLEGSTCGWSCGFATQSSEGPSTPANAALLFLS 120
VGLV L L+ ++S+G++ ++ LE WS + S PS A+A +F++
Sbjct 526 VGLV-KNLALMAYISVGSLPEPILEFLE----EWSMENLEEVS--PSAIADATKIFVN 576
> cpv:cgd2_2140 hypothetical protein
Length=1818
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 58 LMLPFRKTVGLVLLLLLLLLFVSLGTVL 85
+ML FRK +G+ L+ L++L ++LG ++
Sbjct 437 IMLFFRKIIGIFFLINLIILTLALGIII 464
> xla:398890 polr2b, MGC68569; polymerase (RNA) II (DNA directed)
polypeptide B, 140kDa; K03010 DNA-directed RNA polymerase
II subunit RPB2 [EC:2.7.7.6]
Length=949
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 9/78 (11%)
Query 14 HEMRPEDSEALEKLRVQ------QRLCSE--RNYKLQEAEQAHRSAYSLLCRLMLPFRKT 65
H+ R S+ L +L +RL S R+ KL + Q H + + ++C P
Sbjct 460 HQARAGVSQVLNRLTFASTLSHLRRLNSPIGRDGKLAKPRQLHNTLWGMICPAETPEGHA 519
Query 66 VGLVLLLLLLLLFVSLGT 83
VGLV L L+ ++S+G+
Sbjct 520 VGLV-KNLALMAYISVGS 536
> ath:AT5G19310 homeotic gene regulator, putative; K11647 SWI/SNF-related
matrix-associated actin-dependent regulator of chromatin
subfamily A member 2/4 [EC:3.6.4.-]
Length=1064
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 33/117 (28%), Positives = 53/117 (45%), Gaps = 19/117 (16%)
Query 12 GLHEMRPEDSEALEKLRVQQRLCSER-------NYKLQEAEQAHRSA--YSLLCRLMLPF 62
GLH + S++L+ L +Q R C +Y + + + R++ + LL RL LP
Sbjct 649 GLHSGNGK-SKSLQNLTMQLRKCCNHPYLFVGADYNMCKKPEIVRASGKFELLDRL-LPK 706
Query 63 RKTVGLVLLLL--------LLLLFVSLGTVLIRRLEGSTCGWSCGFATQSSEGPSTP 111
K G +LL LL +++SL + RL+GST G + P +P
Sbjct 707 LKKAGHRILLFSQMTRLIDLLEIYLSLNDYMYLRLDGSTKTDQRGILLKQFNEPDSP 763
> mmu:231329 Polr2b, MGC47046, Pol2rb, Rpb140, Rpb2; polymerase
(RNA) II (DNA directed) polypeptide B (EC:2.7.7.6); K03010
DNA-directed RNA polymerase II subunit RPB2 [EC:2.7.7.6]
Length=1174
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 9/78 (11%)
Query 14 HEMRPEDSEALEKLRVQ------QRLCSE--RNYKLQEAEQAHRSAYSLLCRLMLPFRKT 65
H+ R S+ L +L +RL S R+ KL + Q H + + ++C P
Sbjct 460 HQARAGVSQVLNRLTFASTLSHLRRLNSPIGRDGKLAKPRQLHNTLWGMVCPAETPEGHA 519
Query 66 VGLVLLLLLLLLFVSLGT 83
VGLV L L+ ++S+G+
Sbjct 520 VGLV-KNLALMAYISVGS 536
> hsa:5431 POLR2B, POL2RB, RPB2, hRPB140, hsRPB2; polymerase (RNA)
II (DNA directed) polypeptide B, 140kDa (EC:2.7.7.6); K03010
DNA-directed RNA polymerase II subunit RPB2 [EC:2.7.7.6]
Length=1174
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 9/78 (11%)
Query 14 HEMRPEDSEALEKLRVQ------QRLCSE--RNYKLQEAEQAHRSAYSLLCRLMLPFRKT 65
H+ R S+ L +L +RL S R+ KL + Q H + + ++C P
Sbjct 460 HQARAGVSQVLNRLTFASTLSHLRRLNSPIGRDGKLAKPRQLHNTLWGMVCPAETPEGHA 519
Query 66 VGLVLLLLLLLLFVSLGT 83
VGLV L L+ ++S+G+
Sbjct 520 VGLV-KNLALMAYISVGS 536
Lambda K H
0.322 0.136 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2039374804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40