bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6767_orf2
Length=147
Score E
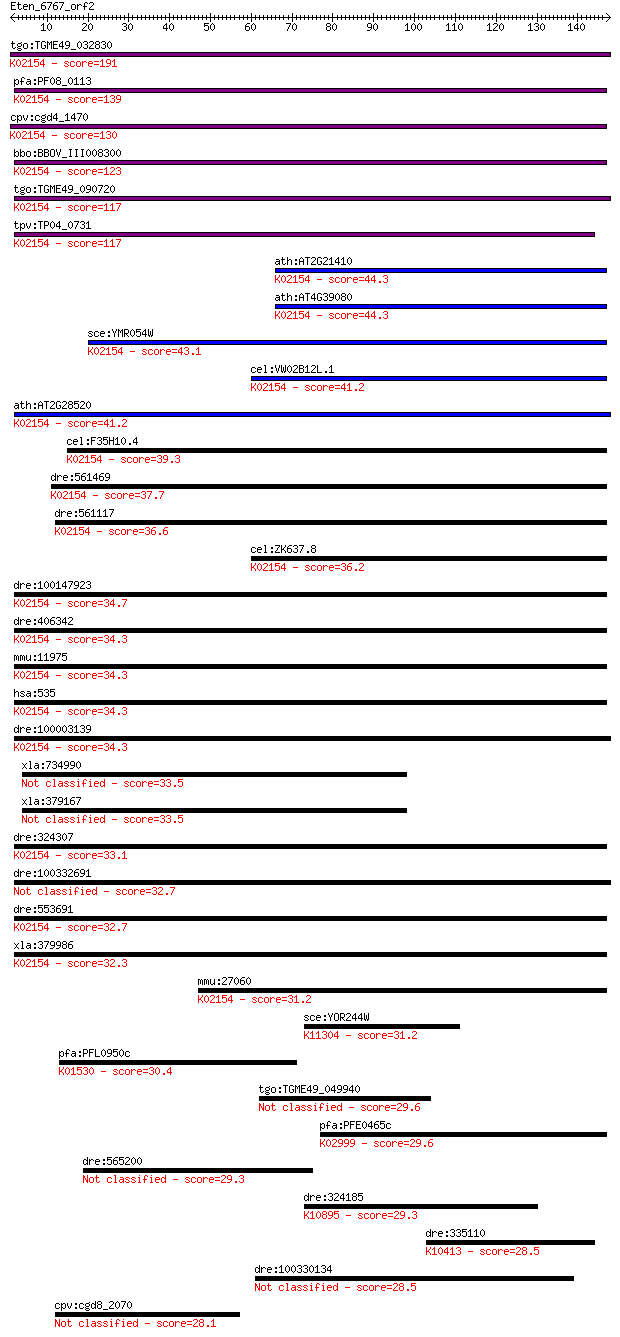
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_032830 vacuolar proton-translocating ATPase subunit... 191 5e-49
pfa:PF08_0113 vacuolar proton translocating ATPase subunit A, ... 139 4e-33
cpv:cgd4_1470 vacuolar proton translocating ATpase with 7 tran... 130 2e-30
bbo:BBOV_III008300 17.m07725; V-type ATPase 116kDa subunit fam... 123 2e-28
tgo:TGME49_090720 vacuolar proton-translocating ATPase subunit... 117 1e-26
tpv:TP04_0731 vacuolar ATP synthase subunit A (EC:3.6.3.14); K... 117 1e-26
ath:AT2G21410 VHA-A2; VHA-A2 (VACUOLAR PROTON ATPASE A2); ATPa... 44.3 1e-04
ath:AT4G39080 VHA-A3; VHA-A3 (VACUOLAR PROTON ATPASE A3); ATPa... 44.3 1e-04
sce:YMR054W STV1; Stv1p (EC:3.6.3.14); K02154 V-type H+-transp... 43.1 3e-04
cel:VW02B12L.1 vha-6; Vacuolar H ATPase family member (vha-6);... 41.2 0.001
ath:AT2G28520 VHA-A1; VHA-A1 (VACUOLAR PROTON ATPASE A 1); ATP... 41.2 0.001
cel:F35H10.4 vha-5; Vacuolar H ATPase family member (vha-5); K... 39.3 0.005
dre:561469 ATPase, H+ transporting, lysosomal V0 subunit a1-li... 37.7 0.011
dre:561117 si:ch211-106a19.2 (EC:3.6.3.6); K02154 V-type H+-tr... 36.6 0.033
cel:ZK637.8 unc-32; UNCoordinated family member (unc-32); K021... 36.2 0.037
dre:100147923 T-cell immune regulator 1-like; K02154 V-type H+... 34.7 0.12
dre:406342 tcirg1, wu:fb44e06, wu:fi46h07; zgc:55891; K02154 V... 34.3 0.13
mmu:11975 Atp6v0a1, AA959968, ATP6a1, Atp6n1, Atp6n1a, Atpv0a1... 34.3 0.13
hsa:535 ATP6V0A1, ATP6N1, ATP6N1A, DKFZp781J1951, Stv1, VPP1, ... 34.3 0.13
dre:100003139 tcirg1, si:dkey-9i23.9; T-cell, immune regulator... 34.3 0.16
xla:734990 rabgap1l, MGC130926, hhl, tbc1d18; RAB GTPase activ... 33.5 0.23
xla:379167 hypothetical protein MGC52980 33.5 0.27
dre:324307 atp6v0a1a, atp6v0a1, wu:fc25b09, zgc:76965; ATPase,... 33.1 0.29
dre:100332691 atp6v0a1; ATPase, H+ transporting, lysosomal V0 ... 32.7 0.37
dre:553691 atp6v0a1b, MGC112214, wu:fj37e01, zgc:112214; ATPas... 32.7 0.43
xla:379986 atp6v0a1, MGC52726; ATPase, H+ transporting, lysoso... 32.3 0.58
mmu:27060 Tcirg1, ATP6N1C, ATP6a3, Atp6i, OC-116, OPTB1, Stv1,... 31.2 1.2
sce:YOR244W ESA1, TAS1; Esa1p (EC:2.3.1.48); K11304 histone ac... 31.2 1.3
pfa:PFL0950c ATPase2; aminophospholipid-transporting P-ATPase ... 30.4 2.2
tgo:TGME49_049940 hypothetical protein 29.6 3.3
pfa:PFE0465c RNA polymerase I; K02999 DNA-directed RNA polymer... 29.6 3.4
dre:565200 stat2; signal transducer and activator of transcrip... 29.3 4.5
dre:324185 fanci, MGC152941, fc21c10, wu:fc21c10, zgc:152941; ... 29.3 5.0
dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic 1... 28.5 8.2
dre:100330134 RETRotransposon-like family member (retr-1)-like 28.5 8.2
cpv:cgd8_2070 membrane associated protein, signal peptide plus... 28.1 9.8
> tgo:TGME49_032830 vacuolar proton-translocating ATPase subunit,
putative (EC:3.6.3.14); K02154 V-type H+-transporting ATPase
subunit I [EC:3.6.3.14]
Length=909
Score = 191 bits (486), Expect = 5e-49, Method: Compositional matrix adjust.
Identities = 91/149 (61%), Positives = 111/149 (74%), Gaps = 2/149 (1%)
Query 1 TKVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREG 60
KV + F YEWP S ++ RL GL ++ DKE+ALAAYE YFL EIS LL+V R G
Sbjct 280 VKVCAAFDAKPYEWPHSAEEAATRLAGLQSLLDDKERALAAYEKYFLSEISLLLEVTRPG 339
Query 61 SSSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSCWIPKKNEEKVRSILR--SMEVTGD 118
SSL+EEW+MFCQKEKA+YA LN F+G+DMTLRC CWIP+ EE++RSIL+ S + G+
Sbjct 340 GSSLLEEWKMFCQKEKAVYATLNQFQGKDMTLRCDCWIPRDKEEEIRSILKDVSTDPNGE 399
Query 119 EQGSAFLLTEKDSAAAMPPTYFKSTEFTD 147
EQ SAFLL EK AMPPTYFK+TEFT+
Sbjct 400 EQASAFLLIEKGQPTAMPPTYFKTTEFTE 428
> pfa:PF08_0113 vacuolar proton translocating ATPase subunit A,
putative; K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=1053
Score = 139 bits (349), Expect = 4e-33, Method: Composition-based stats.
Identities = 66/146 (45%), Positives = 94/146 (64%), Gaps = 3/146 (2%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ + Y+WP + + ++RL+ L ++I DKEKAL AYE YF+ EI L++V
Sbjct 313 KICKAYDVKTYDWPRTYEHAKKRLKELREIINDKEKALKAYEEYFINEIFVLINVVEPNK 372
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSCWIPKKNEEKVRSILRSMEVTGDEQG 121
+SLIEEW++FC+KE+ IY LN FEG D+TLRC CW +EEK+R IL + + ++
Sbjct 373 NSLIEEWKLFCKKERHIYNNLNYFEGSDITLRCDCWYSANDEEKIRHIL--INKSSNDLV 430
Query 122 SAFLLTEKD-SAAAMPPTYFKSTEFT 146
SA LL++K PPTY K+ EFT
Sbjct 431 SALLLSDKILRPNVSPPTYIKTNEFT 456
> cpv:cgd4_1470 vacuolar proton translocating ATpase with 7 transmembrane
regions near C-terminus ; K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=920
Score = 130 bits (327), Expect = 2e-30, Method: Composition-based stats.
Identities = 71/168 (42%), Positives = 90/168 (53%), Gaps = 24/168 (14%)
Query 1 TKVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLD-VPRE 59
+++ F Y WP S + +R+ L +I+DKEKAL AYE Y EI TLL V
Sbjct 278 SRICDAFNVSIYPWPSSYEHAIQRISELNTLIQDKEKALQAYEQYITLEIETLLQPVNSN 337
Query 60 GSSSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSCWIPKKNEEKVRSILRSMEVTGDE 119
+SLIEEWR+FC KEK+IYA LN FEG D+TLR CW P + EEK+R IL + T +
Sbjct 338 NGNSLIEEWRLFCIKEKSIYATLNLFEGSDITLRADCWYPTEEEEKIRKILIAESST--Q 395
Query 120 QGSAFLLTEKDSA---------------------AAMPPTYFKSTEFT 146
AFLLT S + PPTY K+ +FT
Sbjct 396 HVGAFLLTNTSSGGHGVAGIHISEGGSHDDEANISNTPPTYIKTNDFT 443
> bbo:BBOV_III008300 17.m07725; V-type ATPase 116kDa subunit family
protein; K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=927
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 64/153 (41%), Positives = 93/153 (60%), Gaps = 10/153 (6%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ + F + W S +RL+ L ++I+D++KAL A++ YF EI+ LL+ PR
Sbjct 284 KLCNGFQAKTFAWSKSHSHINQRLQELEEIIRDRQKALNAFKRYFREEIACLLECPRPDG 343
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSCWIPKKNEEKVRSILRSMEVTGDEQG 121
+S+IEEW +FC+KEK IY LN FEG D+TLR CW P++ EE +R+ L++ + G +
Sbjct 344 NSVIEEWSLFCRKEKYIYYILNHFEGSDITLRADCWFPEEEEETIRTCLQAEKSEG--RV 401
Query 122 SAFLLTEK--------DSAAAMPPTYFKSTEFT 146
SA LL + D A MPPTY K+ FT
Sbjct 402 SALLLIDHQFKERRYFDDPATMPPTYNKNDVFT 434
> tgo:TGME49_090720 vacuolar proton-translocating ATPase subunit,
putative (EC:3.6.3.14); K02154 V-type H+-transporting ATPase
subunit I [EC:3.6.3.14]
Length=1015
Score = 117 bits (294), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 63/152 (41%), Positives = 89/152 (58%), Gaps = 8/152 (5%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDV----- 56
++ + FG CY WP S ++ ++R ++ ++ DKEK L AYE YFL EIS LL+
Sbjct 328 RLCAAFGARCYSWPGSFEEAEKRFADVSSLLADKEKTLRAYEQYFLSEISILLEPVDADC 387
Query 57 -PREGSSSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSCWIPKKNEEKVRSILRSMEV 115
R LIE WR FC KEKA+YA LN FE D+T+R CW P ++E K+R +L E
Sbjct 388 EGRRRRRPLIEAWRRFCVKEKAVYATLNFFEASDVTIRADCWFPAQDEAKLRVVL--AEQ 445
Query 116 TGDEQGSAFLLTEKDSAAAMPPTYFKSTEFTD 147
+ SAFLL +++ PPT+F+ F +
Sbjct 446 SARSHASAFLLLHPPTSSPSPPTFFRLPPFLE 477
> tpv:TP04_0731 vacuolar ATP synthase subunit A (EC:3.6.3.14);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=936
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 64/157 (40%), Positives = 91/157 (57%), Gaps = 17/157 (10%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ + F + W + + RL+ L DVIKDK++AL AY+ YF GEI+ LL+V R G
Sbjct 280 KLCTGFQAKLFNWCKTQSELAPRLKTLEDVIKDKKRALEAYKDYFRGEIACLLEVIRPGG 339
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSCWIPKKNEEKVRSILRSMEVTGDEQG 121
+S+IEEW +FC+KEK +Y LN FEG D+TLR CW P EEK+R L + + +G
Sbjct 340 NSVIEEWFLFCKKEKYLYYILNHFEGSDITLRADCWFPADEEEKIREHLLAEKASG--SV 397
Query 122 SAFLLTE---------------KDSAAAMPPTYFKST 143
SA LL + ++ + +PPTY K+
Sbjct 398 SALLLVDIQAPFVSVHPSHPGSHENLSHIPPTYNKTN 434
> ath:AT2G21410 VHA-A2; VHA-A2 (VACUOLAR PROTON ATPASE A2); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=821
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 45/86 (52%), Gaps = 10/86 (11%)
Query 66 EEWRMFCQKEKAIYAALNCFEGRDMTLRCSC---WIPKKNEEKVRSILRSMEVTGDEQ-G 121
E+W + +KEKAIY LN D+T +C W P +++ L V + Q G
Sbjct 315 EQWNLKIRKEKAIYHTLNML-SLDVTKKCLVGEGWSPVFAATEIQDALHRAAVDSNSQVG 373
Query 122 SAF-LLTEKDSAAAMPPTYFKSTEFT 146
S F +L K+ MPPT+F++ +FT
Sbjct 374 SIFQVLRTKE----MPPTFFRTNKFT 395
> ath:AT4G39080 VHA-A3; VHA-A3 (VACUOLAR PROTON ATPASE A3); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=821
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 45/86 (52%), Gaps = 10/86 (11%)
Query 66 EEWRMFCQKEKAIYAALNCFEGRDMTLRCSC---WIPKKNEEKVRSILRSMEVTGDEQ-G 121
E W + +KEKAIY LN D+T +C W P +++ L+ V + Q G
Sbjct 314 ELWNLKVRKEKAIYHTLNML-SLDVTKKCLVAEGWSPVFASREIQDALQRAAVDSNSQVG 372
Query 122 SAF-LLTEKDSAAAMPPTYFKSTEFT 146
S F +L K+S PPTYF++ +FT
Sbjct 373 SIFQVLRTKES----PPTYFRTNKFT 394
> sce:YMR054W STV1; Stv1p (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=890
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 32/130 (24%), Positives = 56/130 (43%), Gaps = 16/130 (12%)
Query 20 QTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGSSSLIEEWRMFCQKEKAIY 79
++ E ++ L I D ++ L E E+ + D + W ++EK +Y
Sbjct 317 RSSELVDTLNRQIDDLQRILDTTEQTLHTELLVIHD--------QLPVWSAMTKREKYVY 368
Query 80 AALNCFEGRDMTLRCSCWIPKKNEEKVRSILRS-MEVTGDEQGSAF--LLTEKDSAAAMP 136
LN F+ L W+P ++ L+ +E G E + F +LT K +P
Sbjct 369 TTLNKFQQESQGLIAEGWVPSTELIHLQDSLKDYIETLGSEYSTVFNVILTNK-----LP 423
Query 137 PTYFKSTEFT 146
PTY ++ +FT
Sbjct 424 PTYHRTNKFT 433
> cel:VW02B12L.1 vha-6; Vacuolar H ATPase family member (vha-6);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=865
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 27/92 (29%), Positives = 49/92 (53%), Gaps = 10/92 (10%)
Query 60 GSSSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRC---SCWIPKKNEEKVRSILRSMEVT 116
+++ + +W + K K+I+ LN F D+T +C CW+P+ + +V++ S+ +
Sbjct 296 AAATNLRKWGIMLLKLKSIFHTLNMFS-VDVTQKCLIAECWVPEADIGQVKN---SLHMG 351
Query 117 GDEQGSAF--LLTEKDSAAAMPPTYFKSTEFT 146
GS +L E ++ PPTYFK +FT
Sbjct 352 TIHSGSTVPAILNEMETDK-YPPTYFKLNKFT 382
> ath:AT2G28520 VHA-A1; VHA-A1 (VACUOLAR PROTON ATPASE A 1); ATPase;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=817
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/149 (22%), Positives = 62/149 (41%), Gaps = 14/149 (9%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ FG CY P + ++ + + D E L A + ++++ G
Sbjct 257 KICEAFGANCYPVPEDTTKQRQLTREVLSRLSDLEATLDAGTRHRNNALNSV------GY 310
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSC---WIPKKNEEKVRSILRSMEVTGD 118
S + W ++EKA+Y LN D+T +C W P + ++ +L+ T D
Sbjct 311 S--LTNWITTVRREKAVYDTLNMLN-FDVTKKCLVGEGWCPTFAKTQIHEVLQ--RATFD 365
Query 119 EQGSAFLLTEKDSAAAMPPTYFKSTEFTD 147
++ A PPTYF++ + T+
Sbjct 366 SSSQVGVIFHVMQAVESPPTYFRTNKLTN 394
> cel:F35H10.4 vha-5; Vacuolar H ATPase family member (vha-5);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=873
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 61/137 (44%), Gaps = 18/137 (13%)
Query 15 PLSLKQTQERLEGLADVIKDKEKALA-AYEHYFLGEISTLLDVPREGSSSLIEEWRMFCQ 73
P + K+ Q + I+D + L EH F +L +++ RM
Sbjct 251 PKTFKERQSARNDVRARIQDLQTVLGQTREHRF-----RVLQAAANNHHQWLKQVRMI-- 303
Query 74 KEKAIYAALNCF--EGRDMTLRCSCWIPKKNEEKVRSILRSMEVTGDEQGSAF--LLTEK 129
K ++ LN F +G CWIP K+ E VR +++EV + GS+ +L
Sbjct 304 --KTVFHMLNLFTFDGIGRFFVGECWIPLKHVEDVR---KAIEVGAERSGSSVKPVLNIL 358
Query 130 DSAAAMPPTYFKSTEFT 146
+++ PPTY ++ +FT
Sbjct 359 ETSVT-PPTYNETNKFT 374
> dre:561469 ATPase, H+ transporting, lysosomal V0 subunit a1-like;
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=839
Score = 37.7 bits (86), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 35/138 (25%), Positives = 57/138 (41%), Gaps = 12/138 (8%)
Query 11 CYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGSSSLIEEWRM 70
Y P + ++ + ++ L I+D L E Y L V + S S + W +
Sbjct 249 IYPHPETDEERADVMDSLRTRIQDLHNVLHRTEDY-------LKQVLHKASES-AQSWVL 300
Query 71 FCQKEKAIYAALNC--FEGRDMTLRCSCWIPKKNEEKVRSILRSMEVTGDEQGSAFLLTE 128
+K KAIY LN F+ + L W P + +R L GD +F+
Sbjct 301 QVKKMKAIYHILNLCSFDVTNKCLIAEVWCPVSDLANLRRALEEGSRKGDATVPSFV--N 358
Query 129 KDSAAAMPPTYFKSTEFT 146
+ ++ PPT +S +FT
Sbjct 359 RIPSSDTPPTLLRSNKFT 376
> dre:561117 si:ch211-106a19.2 (EC:3.6.3.6); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=849
Score = 36.6 bits (83), Expect = 0.033, Method: Composition-based stats.
Identities = 36/138 (26%), Positives = 60/138 (43%), Gaps = 14/138 (10%)
Query 12 YEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGSSSLIEEWRMF 71
Y +P S ++ + +EGL I+D L E Y L V + S S + W +
Sbjct 253 YPYPNSNEERTDVVEGLRTRIQDLHTVLHRTEDY-------LRQVLIKASES-VYIWVIQ 304
Query 72 CQKEKAIYAALN--CFEGRDMTLRCSCWIPKKNEEKVRSILRSMEVTGDEQGSAF-LLTE 128
+K KAIY LN F+ + L W P + +R R++E + G+
Sbjct 305 VKKMKAIYHILNLCSFDVTNKCLIAEVWCPVNDLPALR---RALEDGSRKSGATVPSFVN 361
Query 129 KDSAAAMPPTYFKSTEFT 146
+ ++ PPT ++ +FT
Sbjct 362 RIPSSDTPPTLIRTNKFT 379
> cel:ZK637.8 unc-32; UNCoordinated family member (unc-32); K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=899
Score = 36.2 bits (82), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 43/91 (47%), Gaps = 8/91 (8%)
Query 60 GSSSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRC---SCWIPKKNEEKVRSIL-RSMEV 115
+S + W +K K+IY LN F D+T +C W P ++++ L R +
Sbjct 321 AASKNVRMWLTKVRKIKSIYHTLNLFN-IDVTQKCLIAEVWCPIAELDRIKMALKRGTDE 379
Query 116 TGDEQGSAFLLTEKDSAAAMPPTYFKSTEFT 146
+G + S E + A PPTY K+ +FT
Sbjct 380 SGSQVPSILNRMETNEA---PPTYNKTNKFT 407
> dre:100147923 T-cell immune regulator 1-like; K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=793
Score = 34.7 bits (78), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 56/148 (37%), Gaps = 14/148 (9%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ F + +P + + +E L GL I+D + + E Y + L
Sbjct 230 KICDCFHTQTFPYPENQAEREETLNGLRGRIEDIKSVMGETEQYMQQLLVRAL------- 282
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGR--DMTLRCSCWIPKKNEEKVRSILRSMEVTGDE 119
+ + EW + QK KA+ LN D L W P ++S LR G +
Sbjct 283 -ARLPEWVVQVQKCKAVQTVLNLCSPSVTDKCLIAEAWCPVSQLPALQSALRE---GGRK 338
Query 120 QGSAF-LLTEKDSAAAMPPTYFKSTEFT 146
GS + A PPT F + FT
Sbjct 339 SGSNVDSFYNRLPATTSPPTLFPTNSFT 366
> dre:406342 tcirg1, wu:fb44e06, wu:fi46h07; zgc:55891; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=822
Score = 34.3 bits (77), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 56/148 (37%), Gaps = 14/148 (9%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ F + +P + + +E L GL I+D + + E Y + L
Sbjct 230 KICDCFHTQTFPYPENQAEREETLNGLRGRIEDIKSVMGETEQYMQQLLVRAL------- 282
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGR--DMTLRCSCWIPKKNEEKVRSILRSMEVTGDE 119
+ + EW + QK KA+ LN D L W P ++S LR G +
Sbjct 283 -ARLPEWVVQVQKCKAVQTVLNLCSPSVTDKCLIAEAWCPVSQLPALQSALRE---GGRK 338
Query 120 QGSAF-LLTEKDSAAAMPPTYFKSTEFT 146
GS + A PPT F + FT
Sbjct 339 SGSNVDSFYNRLPATTSPPTLFPTNSFT 366
> mmu:11975 Atp6v0a1, AA959968, ATP6a1, Atp6n1, Atp6n1a, Atpv0a1,
Vpp-1, Vpp1; ATPase, H+ transporting, lysosomal V0 subunit
A1 (EC:3.6.3.14); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=838
Score = 34.3 bits (77), Expect = 0.13, Method: Composition-based stats.
Identities = 35/149 (23%), Positives = 62/149 (41%), Gaps = 16/149 (10%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ F Y P + ++ +E G+ I D + L E + + + +
Sbjct 235 KICEGFRASLYPCPETPQERKEMASGVNTRIDDLQMVLNQTEDHRQRVL--------QAA 286
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRC---SCWIPKKNEEKVR-SILRSMEVTG 117
+ I W + +K KAIY LN D+T +C W P + + ++ ++ R E +G
Sbjct 287 AKNIRVWFIKVRKMKAIYHTLN-LCNIDVTQKCLIAEVWCPVTDLDSIQFALRRGTEHSG 345
Query 118 DEQGSAFLLTEKDSAAAMPPTYFKSTEFT 146
S + + PPTY K+ +FT
Sbjct 346 STVPS---ILNRMQTNQTPPTYNKTNKFT 371
> hsa:535 ATP6V0A1, ATP6N1, ATP6N1A, DKFZp781J1951, Stv1, VPP1,
Vph1, a1; ATPase, H+ transporting, lysosomal V0 subunit a1
(EC:3.6.3.14); K02154 V-type H+-transporting ATPase subunit
I [EC:3.6.3.14]
Length=838
Score = 34.3 bits (77), Expect = 0.13, Method: Composition-based stats.
Identities = 35/149 (23%), Positives = 62/149 (41%), Gaps = 16/149 (10%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ F Y P + ++ +E G+ I D + L E + + + +
Sbjct 242 KICEGFRASLYPCPETPQERKEMASGVNTRIDDLQMVLNQTEDHRQRVL--------QAA 293
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRC---SCWIPKKNEEKVR-SILRSMEVTG 117
+ I W + +K KAIY LN D+T +C W P + + ++ ++ R E +G
Sbjct 294 AKNIRVWFIKVRKMKAIYHTLN-LCNIDVTQKCLIAEVWCPVTDLDSIQFALRRGTEHSG 352
Query 118 DEQGSAFLLTEKDSAAAMPPTYFKSTEFT 146
S + + PPTY K+ +FT
Sbjct 353 STVPS---ILNRMQTNQTPPTYNKTNKFT 378
> dre:100003139 tcirg1, si:dkey-9i23.9; T-cell, immune regulator
1, ATPase, H+ transporting, lysosomal V0 subunit A3; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=822
Score = 34.3 bits (77), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 38/149 (25%), Positives = 58/149 (38%), Gaps = 14/149 (9%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEIST-LLDVPREG 60
K+ F + +P L++ + L+GL I D L+ E Y +S + +P
Sbjct 234 KICDCFRTHTFVYPEGLEEREGILQGLESRIVDIRTVLSQTEQYMQQLLSRCVCQMP--- 290
Query 61 SSSLIEEWRMFCQKEKAIYAALNCFEGR--DMTLRCSCWIPKKNEEKVRSILRSMEVTGD 118
+W++ QK KA+ LN D L W P ++S L ME T
Sbjct 291 ------QWKIRVQKCKAVQMVLNLCSPSVTDKCLIAEAWCPVAKLLLLQSAL--MEGTRK 342
Query 119 EQGSAFLLTEKDSAAAMPPTYFKSTEFTD 147
S + A PPT F++ FT
Sbjct 343 SGSSVDSFYNRLPAPTSPPTLFETNAFTS 371
> xla:734990 rabgap1l, MGC130926, hhl, tbc1d18; RAB GTPase activating
protein 1-like
Length=1055
Score = 33.5 bits (75), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 40/96 (41%), Gaps = 10/96 (10%)
Query 4 SSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPR--EGS 61
S F CY + + + +E L +A+A+ + + I+ L VP EGS
Sbjct 124 SVSFHKLCYLGCMKVSAPRNEIEAL--------RAMASMKAQCISPITVTLYVPNIPEGS 175
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSCW 97
+I++ IY L C GRD T C C+
Sbjct 176 VRIIDQTNKSEIASFPIYKVLFCVRGRDGTSECDCF 211
> xla:379167 hypothetical protein MGC52980
Length=1052
Score = 33.5 bits (75), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 40/96 (41%), Gaps = 10/96 (10%)
Query 4 SSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPR--EGS 61
S F CY + + + +E L +A+A+ + + I+ L VP EGS
Sbjct 124 SVSFHKLCYLGCMKVSAPRNEIEAL--------RAMASKKAQCVAPITVTLYVPNIPEGS 175
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSCW 97
+I++ IY L C GRD T C C+
Sbjct 176 VRIIDQTNKSEVASFPIYKVLFCVRGRDGTSECDCF 211
> dre:324307 atp6v0a1a, atp6v0a1, wu:fc25b09, zgc:76965; ATPase,
H+ transporting, lysosomal V0 subunit a isoform 1a (EC:3.6.3.14);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=834
Score = 33.1 bits (74), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 36/150 (24%), Positives = 62/150 (41%), Gaps = 18/150 (12%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ F Y P + ++ +E G+ I D + L E + + + +
Sbjct 235 KICEGFRASLYPCPETPQERKEMAAGVNTRIDDLQMVLNQTEDHRQRVL--------QAA 286
Query 62 SSLIEEWRMFCQKEKAIYAALN-CFEGRDMTLRC---SCWIPKKNEEKVRSILR-SMEVT 116
+ + W + +K KAIY LN C D+T +C W P + + ++ LR E +
Sbjct 287 AKTVRVWFIKVRKMKAIYHTLNLC--NIDVTQKCLIAEIWCPVSDLDSIQFALRRGTERS 344
Query 117 GDEQGSAFLLTEKDSAAAMPPTYFKSTEFT 146
G S + + PPTY K+ +FT
Sbjct 345 GSTVPS---ILNRMQTKQTPPTYNKTNKFT 371
> dre:100332691 atp6v0a1; ATPase, H+ transporting, lysosomal V0
subunit a1
Length=803
Score = 32.7 bits (73), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 37/152 (24%), Positives = 63/152 (41%), Gaps = 20/152 (13%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
KV F Y P +L + +E + ++D L E Y G +S +
Sbjct 247 KVCEGFRASLYSCPKTLYERKEMSNSIMTRMEDLRLVLRRTEEYRAGVLSR--------A 298
Query 62 SSLIEEWRMFCQKEKAIYAALN-CFEGRDMT---LRCSCWIPKKNEEKVRSILRSMEVTG 117
+ ++EW +K KAIY LN C D+T + W P + V++ L + G
Sbjct 299 AEHVQEWGSKVKKMKAIYYTLNLC--NIDITQKLIVAEIWCPVSDLTVVQNAL----IKG 352
Query 118 DEQGSAFL--LTEKDSAAAMPPTYFKSTEFTD 147
EQ + + + + PPT+ ++ FT+
Sbjct 353 SEQSGSSVTPVLNRIQTKQTPPTFNRTNSFTE 384
> dre:553691 atp6v0a1b, MGC112214, wu:fj37e01, zgc:112214; ATPase,
H+ transporting, lysosomal V0 subunit a isoform 1b; K02154
V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=839
Score = 32.7 bits (73), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 36/150 (24%), Positives = 62/150 (41%), Gaps = 18/150 (12%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ F Y P + ++ +E G+ I D + L E + + + +
Sbjct 235 KICEGFRASLYPCPETPQERKEMASGVNTRIDDLQMVLNQTEDHRQRVL--------QAA 286
Query 62 SSLIEEWRMFCQKEKAIYAALN-CFEGRDMTLRC---SCWIPKKNEEKVRSILR-SMEVT 116
+ + W + +K KAIY LN C D+T +C W P + + ++ LR E +
Sbjct 287 AKTMRVWFIKVRKMKAIYHTLNLC--NIDVTQKCLIAEVWCPVSDLDSIQFALRRGTERS 344
Query 117 GDEQGSAFLLTEKDSAAAMPPTYFKSTEFT 146
G S + + PPTY K+ +FT
Sbjct 345 GSTVPS---ILNRMQTKQTPPTYNKTNKFT 371
> xla:379986 atp6v0a1, MGC52726; ATPase, H+ transporting, lysosomal
V0 subunit a1 (EC:3.6.3.14); K02154 V-type H+-transporting
ATPase subunit I [EC:3.6.3.14]
Length=831
Score = 32.3 bits (72), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 36/150 (24%), Positives = 63/150 (42%), Gaps = 18/150 (12%)
Query 2 KVSSVFGGCCYEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGS 61
K+ F Y P + ++ +E G+ I+D + L E + + + +
Sbjct 235 KICEGFRASLYPCPETPQERKEMATGVNTRIEDLQMVLNQTEDHRQRVL--------QAA 286
Query 62 SSLIEEWRMFCQKEKAIYAALN-CFEGRDMTLRC---SCWIPKKNEEKVRSILR-SMEVT 116
+ + W + +K KAIY LN C D+T +C W P + + ++ LR E +
Sbjct 287 AKSLRVWFIKVRKMKAIYHTLNLC--NIDVTQKCLIAEVWCPVADLDSIQFALRRGTEHS 344
Query 117 GDEQGSAFLLTEKDSAAAMPPTYFKSTEFT 146
G S + + PPTY K+ +FT
Sbjct 345 GSTVPS---ILNRMQTNQTPPTYNKTNKFT 371
> mmu:27060 Tcirg1, ATP6N1C, ATP6a3, Atp6i, OC-116, OPTB1, Stv1,
TIRC7, Vph1, oc; T-cell, immune regulator 1, ATPase, H+ transporting,
lysosomal V0 protein A3 (EC:3.6.3.6); K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=834
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 26/104 (25%), Positives = 41/104 (39%), Gaps = 10/104 (9%)
Query 47 LGEISTLLDVPREGSSSLIEEWRMFCQKEKAIYAALN-CFEGRDMTLRC---SCWIPKKN 102
LGE L L+ W++ K KA+Y LN C + T +C W ++
Sbjct 272 LGETDRFLSQVLGRVQQLLPPWQVQIHKMKAVYLTLNQC--SVNTTHKCLIAEVWCAARD 329
Query 103 EEKVRSILRSMEVTGDEQGSAFLLTEKDSAAAMPPTYFKSTEFT 146
V+ L+S G + + + MPPT ++ FT
Sbjct 330 LPTVQQALQS----GSSEEGVSAVAHRIPCQDMPPTLIRTNRFT 369
> sce:YOR244W ESA1, TAS1; Esa1p (EC:2.3.1.48); K11304 histone
acetyltransferase HTATIP [EC:2.3.1.48]
Length=445
Score = 31.2 bits (69), Expect = 1.3, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 23/38 (60%), Gaps = 2/38 (5%)
Query 73 QKEKAIYAALNCFEGRDMTLRCSCWIPKKNEEKVRSIL 110
++E I +N + D+ ++C CW+ K +EE++ IL
Sbjct 6 KEEPGIAKKINSVD--DIIIKCQCWVQKNDEERLAEIL 41
> pfa:PFL0950c ATPase2; aminophospholipid-transporting P-ATPase
(EC:3.6.3.1); K01530 phospholipid-translocating ATPase [EC:3.6.3.1]
Length=1555
Score = 30.4 bits (67), Expect = 2.2, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 31/58 (53%), Gaps = 9/58 (15%)
Query 13 EWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGSSSLIEEWRM 70
E LS+K +E+LE +A+ I++ L I+ + D +EG SS IE+ RM
Sbjct 990 EASLSIKDREEKLESVAEYIEND---------LILQGITGIEDKLQEGVSSTIEDLRM 1038
> tgo:TGME49_049940 hypothetical protein
Length=118
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 62 SSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCSCWIPKKNE 103
+SL+EE F E+A+ +N GR +TL C P E
Sbjct 67 TSLLEEDLQFTSAEEALSLTVNTLFGRGLTLEKRCLTPTTQE 108
> pfa:PFE0465c RNA polymerase I; K02999 DNA-directed RNA polymerase
I subunit RPA1 [EC:2.7.7.6]
Length=2914
Score = 29.6 bits (65), Expect = 3.4, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 36/72 (50%), Gaps = 3/72 (4%)
Query 77 AIYAALNCFEGRDMTLRCSCWIPKKNEEKVRSILRSMEVTGDEQGSAFL--LTEKDSAAA 134
++Y+ + +D+ ++ KNEE + IL + DE+ + FL +T K A
Sbjct 1418 SLYSCVQYVLNKDLFNLYEKYLNGKNEEANKHILYN-NCQIDERNNHFLNSVTNKLYKAL 1476
Query 135 MPPTYFKSTEFT 146
PT+F ST FT
Sbjct 1477 KEPTFFNSTSFT 1488
> dre:565200 stat2; signal transducer and activator of transcription
2
Length=835
Score = 29.3 bits (64), Expect = 4.5, Method: Composition-based stats.
Identities = 22/60 (36%), Positives = 30/60 (50%), Gaps = 8/60 (13%)
Query 19 KQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDVPREGSSSLIEE----WRMFCQK 74
++ +LEGL ++ AL FL EIS +LDV SSLI+E W+ QK
Sbjct 187 EEKNRQLEGLQKMLN----ALDKSRKDFLSEISAMLDVADTLGSSLIDEELVDWKRRQQK 242
> dre:324185 fanci, MGC152941, fc21c10, wu:fc21c10, zgc:152941;
Fanconi anemia, complementation group I; K10895 fanconi anemia
group I protein
Length=1327
Score = 29.3 bits (64), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 31/57 (54%), Gaps = 3/57 (5%)
Query 73 QKEKAIYAALNCFEGRDMTLRCSCWIPKKNEEKVRSILRSMEVTGDEQGSAFLLTEK 129
+KEK +L C EG LR + ++ +V + L S++V+G+ +G LTE+
Sbjct 901 KKEKGQSVSLLCLEG---LLRVFTTVLQRYPTRVSNFLSSLDVSGEGEGDKSDLTEQ 954
> dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4643
Score = 28.5 bits (62), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query 103 EEKVRSILRSMEVTGDEQGSAFLLTEKDSAAAMPPTYFKST 143
E++ + + ++ E G+E S L ++D+AA + P YF ST
Sbjct 2390 EDEAQRMRKATEDEGEENASPMLQIQRDAAAVLQP-YFTST 2429
> dre:100330134 RETRotransposon-like family member (retr-1)-like
Length=1075
Score = 28.5 bits (62), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 35/81 (43%), Gaps = 9/81 (11%)
Query 61 SSSLIEEWRMFCQKEKAIYAALNCFEGRDMTLRCS---CWIPKKNEEKVRSILRSMEVTG 117
SS+L + WR F Q + ++A G D +CS WI EK R I + +T
Sbjct 15 SSNLPDAWRKFRQHAELMFAGPLKKRGEDE--KCSYLLLWI----GEKGRDIFNTWALTA 68
Query 118 DEQGSAFLLTEKDSAAAMPPT 138
DE +K A MP T
Sbjct 69 DEAKVLQTYYDKYEAYVMPKT 89
> cpv:cgd8_2070 membrane associated protein, signal peptide plus
transmembrane domain or GPI anchor at
Length=793
Score = 28.1 bits (61), Expect = 9.8, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 28/45 (62%), Gaps = 3/45 (6%)
Query 12 YEWPLSLKQTQERLEGLADVIKDKEKALAAYEHYFLGEISTLLDV 56
Y++P+SLK + L L++ +K EK+L + +G+IST + V
Sbjct 607 YKYPVSLKVNLKMLVNLSETLKIGEKSLGDIK---VGDISTCIKV 648
Lambda K H
0.317 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40