bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6741_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
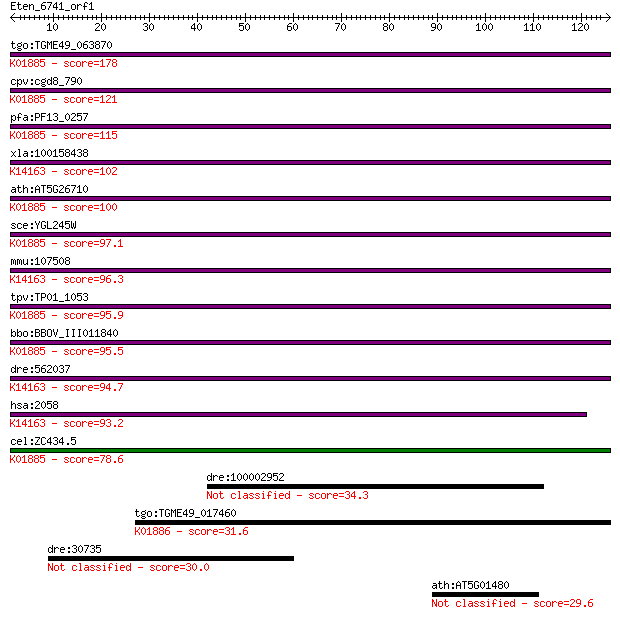
tgo:TGME49_063870 glutamyl-tRNA synthetase, putative (EC:6.1.1... 178 4e-45
cpv:cgd8_790 glutamate--tRNA ligase ; K01885 glutamyl-tRNA syn... 121 7e-28
pfa:PF13_0257 glutamate--tRNA ligase, putative (EC:6.1.1.17); ... 115 2e-26
xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163 bi... 102 4e-22
ath:AT5G26710 glutamate-tRNA ligase, putative / glutamyl-tRNA ... 100 2e-21
sce:YGL245W GUS1, GSN1; GluRS (EC:6.1.1.17); K01885 glutamyl-t... 97.1 1e-20
mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs; g... 96.3 2e-20
tpv:TP01_1053 glutamyl-tRNA synthetase; K01885 glutamyl-tRNA s... 95.9 3e-20
bbo:BBOV_III011840 17.m08008; glutamyl-tRNA synthetase family ... 95.5 4e-20
dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA ... 94.7 6e-20
hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;... 93.2 2e-19
cel:ZC434.5 ers-2; glutamyl(E)/glutaminyl(Q) tRNA Synthetase f... 78.6 4e-15
dre:100002952 reverse transcriptase/ribonuclease H/putative me... 34.3 0.096
tgo:TGME49_017460 glutaminyl-tRNA synthetase, putative (EC:6.1... 31.6 0.74
dre:30735 irbp; interphotoreceptor retinoid-binding protein 30.0 2.1
ath:AT5G01480 DC1 domain-containing protein 29.6 2.4
> tgo:TGME49_063870 glutamyl-tRNA synthetase, putative (EC:6.1.1.18);
K01885 glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=786
Score = 178 bits (451), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 78/125 (62%), Positives = 102/125 (81%), Gaps = 1/125 (0%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDKLWT N+Q+IDPIVPRFMAV + D V V I GAPE V ++ RR+HAKNE+LG+ L
Sbjct 534 EWDKLWTKNKQIIDPIVPRFMAVGK-DAVPVCIKGAPETVESKKRRMHAKNESLGEADLL 592
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKK 120
L N + I+ DDA LC+DGEEVTLMHWGNC+F+++ + ++G+++ I A LHLEGDF+KTKK
Sbjct 593 LFNKVFIDRDDAALCEDGEEVTLMHWGNCIFDKVVKTASGEISEIQATLHLEGDFRKTKK 652
Query 121 KLNWV 125
KL+W+
Sbjct 653 KLHWL 657
> cpv:cgd8_790 glutamate--tRNA ligase ; K01885 glutamyl-tRNA synthetase
[EC:6.1.1.17]
Length=555
Score = 121 bits (303), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 57/125 (45%), Positives = 81/125 (64%), Gaps = 1/125 (0%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDKLWT+N+Q++DP+VPRF AV + D V + +S APE R LH KN LG+ +
Sbjct 320 EWDKLWTINKQLMDPVVPRFFAVGQ-DAVALNLSEAPEEPLINQRDLHQKNPDLGKGQIV 378
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKK 120
++ +ILI+ DDA GEE+TLM WGN + + + + KL+LEGDF+ TKK
Sbjct 379 MYKSILIDRDDATQILSGEEITLMKWGNAIIQDISQIDKQPTGIFEGKLNLEGDFRSTKK 438
Query 121 KLNWV 125
K++W+
Sbjct 439 KIHWL 443
> pfa:PF13_0257 glutamate--tRNA ligase, putative (EC:6.1.1.17);
K01885 glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=863
Score = 115 bits (289), Expect = 2e-26, Method: Composition-based stats.
Identities = 51/125 (40%), Positives = 85/125 (68%), Gaps = 1/125 (0%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
+WDKLW++N+Q+IDPI+PR+ AV + + + ++ + V + R LH KN++LG +Y
Sbjct 602 QWDKLWSINKQIIDPIIPRYAAVDKNSSILLILTDLTDQVIQKERDLHMKNKSLGTCNMY 661
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKK 120
+N LIE +DAQ + EE+TL+ GN + + +E+++ GK+ I+A + GDFK TKK
Sbjct 662 YNNKYLIELEDAQTLLENEEITLIKLGNIIIKNIEKEN-GKIKQINALSNFHGDFKTTKK 720
Query 121 KLNWV 125
K++W+
Sbjct 721 KIHWL 725
> xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17 6.1.1.15]
Length=1499
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 51/126 (40%), Positives = 78/126 (61%), Gaps = 4/126 (3%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDK+W N++VIDP+ PR+ A+ R V V I A E + + H KN +G P++
Sbjct 479 EWDKIWAFNKKVIDPVAPRYTALLRNQVVPVNIPDAQEGMLEVAK--HPKNTEVGLKPVW 536
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEG-DFKKTK 119
+LIE DA+ +GE VT ++WGN + ++ RD +GK+ ++ AKL+LE D+KKT
Sbjct 537 YGPKVLIEGADAETLSEGETVTFINWGNLIITKIHRDQSGKIQSLDAKLNLENKDYKKT- 595
Query 120 KKLNWV 125
K+ W+
Sbjct 596 TKITWL 601
> ath:AT5G26710 glutamate-tRNA ligase, putative / glutamyl-tRNA
synthetase, putatuve / GluRS, putative (EC:6.1.1.17); K01885
glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=719
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 50/125 (40%), Positives = 72/125 (57%), Gaps = 0/125 (0%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDKLW++N+++IDP+ PR AV V T++ P+ + H K E G+
Sbjct 504 EWDKLWSINKRIIDPVCPRHTAVVAERRVLFTLTDGPDEPFVRMIPKHKKFEGAGEKATT 563
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKK 120
+I +E DA GEEVTLM WGN + + + +D G+VTA+S L+L+G K TK
Sbjct 564 FTKSIWLEEADASAISVGEEVTLMDWGNAIVKEITKDEEGRVTALSGVLNLQGSVKTTKL 623
Query 121 KLNWV 125
KL W+
Sbjct 624 KLTWL 628
> sce:YGL245W GUS1, GSN1; GluRS (EC:6.1.1.17); K01885 glutamyl-tRNA
synthetase [EC:6.1.1.17]
Length=708
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 49/125 (39%), Positives = 73/125 (58%), Gaps = 5/125 (4%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EW+ +W N++VIDPI PR A+ + + S AP+ + + H KN A+G+ +
Sbjct 493 EWNLIWAFNKKVIDPIAPRHTAIVNPVKIHLEGSEAPQEPKIEMKPKHKKNPAVGEKKVI 552
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKK 120
+ I+++ DDA + EEVTLM WGN + + D + + AKL+LEGDFKKTK
Sbjct 553 YYKDIVVDKDDADVINVDEEVTLMDWGNVIITKKNDDGS-----MVAKLNLEGDFKKTKH 607
Query 121 KLNWV 125
KL W+
Sbjct 608 KLTWL 612
> mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 48/126 (38%), Positives = 79/126 (62%), Gaps = 4/126 (3%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDK+W N++VIDP+ PR++A+ + + V V + A E + R H KN +G P++
Sbjct 488 EWDKIWAFNKKVIDPVAPRYVALLKKEVVPVNVLDAQEEMKEVAR--HPKNPDVGLKPVW 545
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEG-DFKKTK 119
+ IE DA+ +GE VT ++WGN ++ +++ GK+T++ AKL+LE D+KKT
Sbjct 546 YSPKVFIEGADAETFSEGEMVTFINWGNINITKIHKNADGKITSLDAKLNLENKDYKKT- 604
Query 120 KKLNWV 125
K+ W+
Sbjct 605 TKITWL 610
> tpv:TP01_1053 glutamyl-tRNA synthetase; K01885 glutamyl-tRNA
synthetase [EC:6.1.1.17]
Length=530
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 53/125 (42%), Positives = 74/125 (59%), Gaps = 11/125 (8%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDKLW N+Q+IDP PR+ AV +D V + + E AT+ R LH KN L ++ L
Sbjct 350 EWDKLWAKNKQIIDPESPRYTAVV-SDHVVLKVVNY-EEPATKTRPLHPKNPDLREIDLV 407
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKK 120
+ ++IE DD L + EEVTLM WGN ++ + KL+L+GDFK TKK
Sbjct 408 FGDQVMIERDDFNLIEKSEEVTLMKWGNAFVDK---------ANLQLKLNLQGDFKLTKK 458
Query 121 KLNWV 125
K++W+
Sbjct 459 KIHWL 463
> bbo:BBOV_III011840 17.m08008; glutamyl-tRNA synthetase family
protein (EC:6.1.1.18); K01885 glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=730
Score = 95.5 bits (236), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 51/126 (40%), Positives = 74/126 (58%), Gaps = 11/126 (8%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPE-NVATQPRRLHAKNEALGQVPL 59
EWDKLW N+Q+IDPIVPR+ AV D V++ + + + R LH K+ +G+ +
Sbjct 519 EWDKLWAKNKQIIDPIVPRYAAVE-IDAVELKLDNFSQVELPPSKRMLHPKDPNMGECDI 577
Query 60 YLHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTK 119
+ T+L++ DA DGEEVTLM WGN + A + +++ GDFKKTK
Sbjct 578 WFTPTVLLDRVDADEIVDGEEVTLMRWGNVFVSK---------PAFTGQINPGGDFKKTK 628
Query 120 KKLNWV 125
KKL+W+
Sbjct 629 KKLHWL 634
> dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA
synthetase; K14163 bifunctional glutamyl/prolyl-tRNA synthetase
[EC:6.1.1.17 6.1.1.15]
Length=1511
Score = 94.7 bits (234), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 47/126 (37%), Positives = 76/126 (60%), Gaps = 4/126 (3%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDK+W N++VIDP+ PRF A+ + + V +S A E + P+ H KN +G ++
Sbjct 487 EWDKIWAFNKKVIDPVAPRFTALLSSQVIPVCVSEAKETMKEVPK--HPKNADVGLKQVW 544
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEG-DFKKTK 119
+ IE DA+ +GE VT ++WGN + ++ RDS+G + ++ L+LE D+KKT
Sbjct 545 YGPKVFIEGADAETFTEGEIVTFINWGNIIITKIHRDSSGAILSLDGHLNLENTDYKKT- 603
Query 120 KKLNWV 125
K+ W+
Sbjct 604 TKITWL 609
> hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 44/121 (36%), Positives = 75/121 (61%), Gaps = 3/121 (2%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDK+W N++VIDP+ PR++A+ + + + V + A E + + H KN +G P++
Sbjct 488 EWDKIWAFNKKVIDPVAPRYVALLKKEVIPVNVPEAQEEMKEVAK--HPKNPEVGLKPVW 545
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEG-DFKKTK 119
+ IE DA+ +GE VT ++WGN ++ +++ GK+ ++ AKL+LE D+KKT
Sbjct 546 YSPKVFIEGADAETFSEGEMVTFINWGNLNITKIHKNADGKIISLDAKLNLENKDYKKTT 605
Query 120 K 120
K
Sbjct 606 K 606
> cel:ZC434.5 ers-2; glutamyl(E)/glutaminyl(Q) tRNA Synthetase
family member (ers-2); K01885 glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=1149
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 48/131 (36%), Positives = 75/131 (57%), Gaps = 13/131 (9%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTD---GVKVT--ISGAPENVATQPRRLHAKNEALG 55
EWDK+W N++VIDP+ PR+ A+ T +++T IS NV+ LH KN +G
Sbjct 484 EWDKIWAFNKKVIDPVAPRYTALDSTSPLVSIELTDSISDDTSNVS-----LHPKNAEIG 538
Query 56 QVPLYLHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEG-D 114
++ +L+E DA ++GE VT ++WGN ++E+ +T ISA L L+ D
Sbjct 539 SKDVHKGKKLLLEQVDAAALKEGEIVTFVNWGNIKIGKIEKKG-AVITKISATLQLDNTD 597
Query 115 FKKTKKKLNWV 125
+KKT K+ W+
Sbjct 598 YKKT-TKVTWL 607
> dre:100002952 reverse transcriptase/ribonuclease H/putative
methyltransferase-like
Length=1603
Score = 34.3 bits (77), Expect = 0.096, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 35/71 (49%), Gaps = 5/71 (7%)
Query 42 TQPRRLHAKNEALGQVPLYLHNTILIETDDAQL-CQDGEEVTLMHWGNCVFERLERDSTG 100
T PR L Q L +L ETD +L C + EE+ + CVF + ER+S
Sbjct 1323 TSPRSDTVTIRGLSQARLSASALVLQETDTVELSCNNTEELKMEM---CVFNKYERESDS 1379
Query 101 KVTAISAKLHL 111
KV++ S +L L
Sbjct 1380 KVSS-SCQLSL 1389
> tgo:TGME49_017460 glutaminyl-tRNA synthetase, putative (EC:6.1.1.18);
K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=860
Score = 31.6 bits (70), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 50/116 (43%), Gaps = 19/116 (16%)
Query 27 DGVKVTISGAPENVATQPRRLHAKNEALGQVPLYLHNTILIETDD--------AQLCQDG 78
D + VTI+ + V T H ++E+ G L+ + I+ DD + G
Sbjct 593 DPIAVTITNYGDKVETITAANHPEDESFGTRELHFSKKLYIDRDDFMENPPAGYRRLAPG 652
Query 79 EEVTLMH--WGNCVFERLERDSTGKVTAI-------SAKLHLEGDFKKTKKKLNWV 125
EV L H W CV + +D++G VT + + + D +K K ++W+
Sbjct 653 AEVRLKHAYWIKCV--DVVKDASGLVTELLCTYDPQTKNCSVAPDGRKVKGAIHWL 706
> dre:30735 irbp; interphotoreceptor retinoid-binding protein
Length=615
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 9 NRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPL 59
N +++ P +A TDGVK TIS + V +P + A A+ +PL
Sbjct 57 NTEILSISDPTMLANVLTDGVKKTISDSRVKVTYEPDLILAAPPAMPDIPL 107
> ath:AT5G01480 DC1 domain-containing protein
Length=413
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 89 CVFERLERDSTGKVTAISAKLH 110
C ER+++D GKVT IS +H
Sbjct 14 CPVERIQKDKDGKVTTISGGIH 35
Lambda K H
0.318 0.134 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064871684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40