bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6726_orf2
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
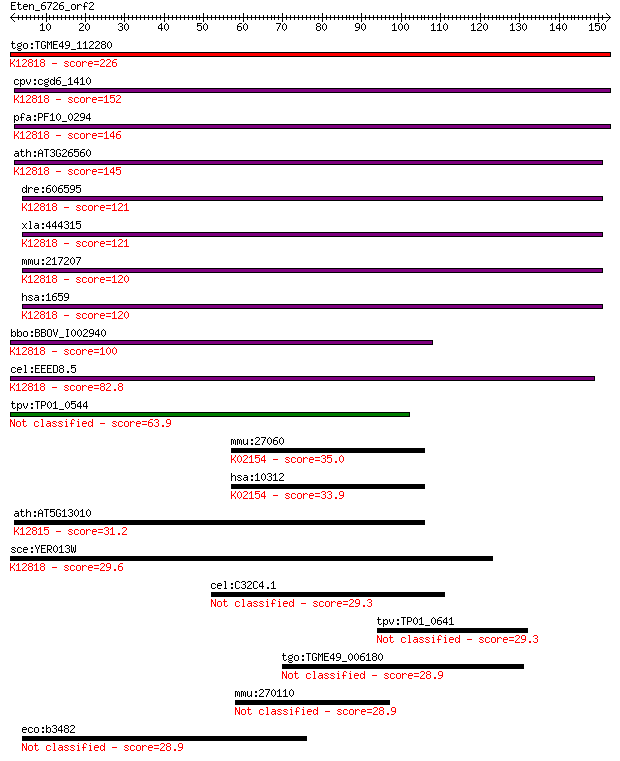
tgo:TGME49_112280 ATP-dependent RNA helicase, putative (EC:3.4... 226 2e-59
cpv:cgd6_1410 pre-mRNA splicing factor ATP-dependent RNA helic... 152 3e-37
pfa:PF10_0294 RNA helicase, putative; K12818 ATP-dependent RNA... 146 3e-35
ath:AT3G26560 ATP-dependent RNA helicase, putative; K12818 ATP... 145 7e-35
dre:606595 im:7153552; K12818 ATP-dependent RNA helicase DHX8/... 121 7e-28
xla:444315 MGC80994 protein; K12818 ATP-dependent RNA helicase... 121 8e-28
mmu:217207 Dhx8, Ddx8, KIAA4096, MGC31290, mDEAH6, mKIAA4096; ... 120 2e-27
hsa:1659 DHX8, DDX8, HRH1, PRP22, PRPF22; DEAH (Asp-Glu-Ala-Hi... 120 2e-27
bbo:BBOV_I002940 19.m02117; RNA helicase; K12818 ATP-dependent... 100 3e-21
cel:EEED8.5 mog-5; Masculinisation Of Germline family member (... 82.8 3e-16
tpv:TP01_0544 RNA helicase 63.9 2e-10
mmu:27060 Tcirg1, ATP6N1C, ATP6a3, Atp6i, OC-116, OPTB1, Stv1,... 35.0 0.090
hsa:10312 TCIRG1, ATP6N1C, ATP6V0A3, Atp6i, OC-116kDa, OC116, ... 33.9 0.21
ath:AT5G13010 EMB3011 (embryo defective 3011); ATP binding / R... 31.2 1.3
sce:YER013W PRP22; DEAH-box RNA-dependent ATPase/ATP-dependent... 29.6 4.0
cel:C32C4.1 hypothetical protein 29.3 4.9
tpv:TP01_0641 RNA helicase 29.3 5.3
tgo:TGME49_006180 hypothetical protein 28.9 6.7
mmu:270110 Irf2bp2, E130305N23Rik; interferon regulatory facto... 28.9 6.8
eco:b3482 rhsB, ECK3466, JW5679; RhsB element core protein RshB 28.9 7.6
> tgo:TGME49_112280 ATP-dependent RNA helicase, putative (EC:3.4.22.44);
K12818 ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1206
Score = 226 bits (577), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 117/152 (76%), Positives = 135/152 (88%), Gaps = 1/152 (0%)
Query 1 RKLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILPAQEAEEDVEIEVVDEEPLFIRG 60
RKLMSD+DKWEAQQL SGL++ +E+P +DEE G ILP+ E +EDVE+E+ ++E LF+RG
Sbjct 379 RKLMSDFDKWEAQQLLHSGLLTREEHPLFDEELG-ILPSAEVDEDVEVEIREDEALFLRG 437
Query 61 QTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWE 120
QTTR GMQLSPVKIVANPDGSLARAAATA LAKERR++R AQEAAILDSIPKDMSRPWE
Sbjct 438 QTTRTGMQLSPVKIVANPDGSLARAAATATALAKERREIRNAQEAAILDSIPKDMSRPWE 497
Query 121 DPNPQPGERTIAQALQGLGQTGYEMPEWKRLY 152
DP P PGERTIAQAL+GLGQT YEMPEWK++Y
Sbjct 498 DPAPGPGERTIAQALKGLGQTSYEMPEWKKMY 529
> cpv:cgd6_1410 pre-mRNA splicing factor ATP-dependent RNA helicase
; K12818 ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1005
Score = 152 bits (385), Expect = 3e-37, Method: Composition-based stats.
Identities = 73/152 (48%), Positives = 102/152 (67%), Gaps = 1/152 (0%)
Query 2 KLMSDYDKWEAQQLQRSGLVSSKENPY-YDEEAGGILPAQEAEEDVEIEVVDEEPLFIRG 60
++ +DY+KWE QL SG++S E PY + G + Q E EIE+ + EPLF+RG
Sbjct 176 RVSNDYEKWEIMQLLNSGVISRDEIPYDICDTTGDTIDFQNVEISTEIELRNYEPLFLRG 235
Query 61 QTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWE 120
Q+ + S +++V NP+GSL +AA A +A+ERR++R QE ++DSIP+DM+RPWE
Sbjct 236 QSIKKFNFDSSIQVVVNPEGSLNKAAELASNIARERREIRDFQEKTLIDSIPRDMNRPWE 295
Query 121 DPNPQPGERTIAQALQGLGQTGYEMPEWKRLY 152
DPNP+ GERTIA AL+G+G PEWKR Y
Sbjct 296 DPNPEAGERTIASALRGIGMNSQTTPEWKRQY 327
> pfa:PF10_0294 RNA helicase, putative; K12818 ATP-dependent RNA
helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1290
Score = 146 bits (368), Expect = 3e-35, Method: Composition-based stats.
Identities = 74/151 (49%), Positives = 108/151 (71%), Gaps = 1/151 (0%)
Query 2 KLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILPAQEAEEDVEIEVVDEEPLFIRGQ 61
K+ SDY KWE QQL +SG+V + + ++ E+ +EIEV ++EP F++GQ
Sbjct 460 KIQSDYSKWEIQQLIKSGVVFDENIRNEYKNLKIDEKIEDEEDIIEIEVNEKEPAFLKGQ 519
Query 62 TTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWED 121
TT+AG +LSP++I+ N +GSLA+A T LAKER++ +Q ++ AI D+IPKD+SRPWED
Sbjct 520 TTKAGAKLSPIQIIVNAEGSLAKAITTTSALAKERKEQKQNEQNAIYDNIPKDISRPWED 579
Query 122 PNPQPGERTIAQALQGLGQTGYEMPEWKRLY 152
P P GERTIA+AL+ +G+ Y++PEWK+ Y
Sbjct 580 PKPNLGERTIAEALKNIGKN-YDIPEWKKNY 609
> ath:AT3G26560 ATP-dependent RNA helicase, putative; K12818 ATP-dependent
RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1168
Score = 145 bits (365), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 79/150 (52%), Positives = 112/150 (74%), Gaps = 2/150 (1%)
Query 2 KLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGIL-PAQEAEEDVEIEVVDEEPLFIRG 60
K MS ++WEA+QL SG++ E P YDE+ G+L + AEE++EIE+ ++EP F++G
Sbjct 343 KKMSSPERWEAKQLIASGVLRVDEFPMYDEDGDGMLYQEEGAEEELEIEMNEDEPAFLQG 402
Query 61 QTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWE 120
QT R + +SPVKI NP+GSL+RAAA L KERR++R+ Q+ +LDSIPKD++RPWE
Sbjct 403 QT-RYSVDMSPVKIFKNPEGSLSRAAALQSALTKERREMREQQQRTMLDSIPKDLNRPWE 461
Query 121 DPNPQPGERTIAQALQGLGQTGYEMPEWKR 150
DP P+ GER +AQ L+G+G + Y+MPEWK+
Sbjct 462 DPMPETGERHLAQELRGVGLSAYDMPEWKK 491
> dre:606595 im:7153552; K12818 ATP-dependent RNA helicase DHX8/PRP22
[EC:3.6.4.13]
Length=1210
Score = 121 bits (304), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 73/149 (48%), Positives = 103/149 (69%), Gaps = 4/149 (2%)
Query 4 MSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILP--AQEAEEDVEIEVVDEEPLFIRGQ 61
+SD +KWE +Q+ + ++S +E P +DEE G ILP E +ED+EIE+V+EEP F+RG
Sbjct 384 ISDPEKWEIKQMIAANVLSKEEFPDFDEETG-ILPKVDDEEDEDLEIELVEEEPPFLRGH 442
Query 62 TTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWED 121
T + M +SPVKIV NPDGSL++AA LAKERR+V+QAQ A +DSIP +++ W D
Sbjct 443 T-KQSMDMSPVKIVKNPDGSLSQAAMMQSALAKERREVKQAQREAEMDSIPMGLNKHWVD 501
Query 122 PNPQPGERTIAQALQGLGQTGYEMPEWKR 150
P P R IA ++G+G ++PEWK+
Sbjct 502 PLPDVDGRQIAANMRGIGMMPNDIPEWKK 530
> xla:444315 MGC80994 protein; K12818 ATP-dependent RNA helicase
DHX8/PRP22 [EC:3.6.4.13]
Length=793
Score = 121 bits (304), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 72/149 (48%), Positives = 103/149 (69%), Gaps = 4/149 (2%)
Query 4 MSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILP--AQEAEEDVEIEVVDEEPLFIRGQ 61
+SD +KWE +Q+ + ++S +E P +DEE G ILP E +ED+EIE+V+EEP F+RG
Sbjct 350 ISDPEKWEIKQMIAANVLSKEEFPDFDEETG-ILPKVDDEEDEDLEIELVEEEPPFLRGH 408
Query 62 TTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWED 121
T + M +SP+KIV NPDGSL++AA LAKERR+V+QAQ A +DSIP +++ W D
Sbjct 409 T-KQSMDMSPIKIVKNPDGSLSQAAMMQSALAKERREVKQAQREAEMDSIPMGLNKHWVD 467
Query 122 PNPQPGERTIAQALQGLGQTGYEMPEWKR 150
P P R IA ++G+G ++PEWK+
Sbjct 468 PLPDVDGRQIAANMRGIGMMPNDIPEWKK 496
> mmu:217207 Dhx8, Ddx8, KIAA4096, MGC31290, mDEAH6, mKIAA4096;
DEAH (Asp-Glu-Ala-His) box polypeptide 8 (EC:3.6.4.13); K12818
ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1244
Score = 120 bits (300), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 71/149 (47%), Positives = 103/149 (69%), Gaps = 4/149 (2%)
Query 4 MSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILP--AQEAEEDVEIEVVDEEPLFIRGQ 61
+SD +KWE +Q+ + ++S +E P +DEE G ILP E +ED+EIE+V+EEP F+RG
Sbjct 418 ISDPEKWEIKQMIAANVLSKEEFPDFDEETG-ILPKVDDEEDEDLEIELVEEEPPFLRGH 476
Query 62 TTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWED 121
T + M +SP+KIV NPDGSL++AA LAKERR+++QAQ A +DSIP +++ W D
Sbjct 477 T-KQSMDMSPIKIVKNPDGSLSQAAMMQSALAKERRELKQAQREAEMDSIPMGLNKHWVD 535
Query 122 PNPQPGERTIAQALQGLGQTGYEMPEWKR 150
P P R IA ++G+G ++PEWK+
Sbjct 536 PLPDAEGRQIAANMRGIGMMPNDIPEWKK 564
> hsa:1659 DHX8, DDX8, HRH1, PRP22, PRPF22; DEAH (Asp-Glu-Ala-His)
box polypeptide 8 (EC:3.6.4.13); K12818 ATP-dependent RNA
helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1220
Score = 120 bits (300), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 71/149 (47%), Positives = 103/149 (69%), Gaps = 4/149 (2%)
Query 4 MSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILP--AQEAEEDVEIEVVDEEPLFIRGQ 61
+SD +KWE +Q+ + ++S +E P +DEE G ILP E +ED+EIE+V+EEP F+RG
Sbjct 394 ISDPEKWEIKQMIAANVLSKEEFPDFDEETG-ILPKVDDEEDEDLEIELVEEEPPFLRGH 452
Query 62 TTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWED 121
T + M +SP+KIV NPDGSL++AA LAKERR+++QAQ A +DSIP +++ W D
Sbjct 453 T-KQSMDMSPIKIVKNPDGSLSQAAMMQSALAKERRELKQAQREAEMDSIPMGLNKHWVD 511
Query 122 PNPQPGERTIAQALQGLGQTGYEMPEWKR 150
P P R IA ++G+G ++PEWK+
Sbjct 512 PLPDAEGRQIAANMRGIGMMPNDIPEWKK 540
> bbo:BBOV_I002940 19.m02117; RNA helicase; K12818 ATP-dependent
RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1156
Score = 100 bits (248), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 51/110 (46%), Positives = 75/110 (68%), Gaps = 9/110 (8%)
Query 1 RKLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILPAQ---EAEEDVEIEVVDEEPLF 57
R++M+D ++WE QQL SG++ E D L AQ E +E+++IE+ D P F
Sbjct 336 RRMMTDLERWEHQQLVNSGVLPKSERVAMD------LAAQHEPELDEEIDIEINDACPTF 389
Query 58 IRGQTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAI 107
++GQT R+G++LSP+KIV+NP+GSLAR AT+ T+AKERR+ + QE I
Sbjct 390 LKGQTRRSGIELSPIKIVSNPEGSLARTIATSSTIAKERRETERMQEDTI 439
> cel:EEED8.5 mog-5; Masculinisation Of Germline family member
(mog-5); K12818 ATP-dependent RNA helicase DHX8/PRP22 [EC:3.6.4.13]
Length=1200
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 57/155 (36%), Positives = 92/155 (59%), Gaps = 10/155 (6%)
Query 1 RKLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILPAQEAE---EDVEIEVVDEEPLF 57
R +S ++WE +Q+Q +G++++ + P +DEE G +L + E ED+EIE+V++EP F
Sbjct 362 RVRISTPERWELRQMQGAGVLTATDMPDFDEEMG-VLRNYDDESDGEDIEIELVEDEPDF 420
Query 58 IRGQTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVR-QAQEAAILDSIPKDMS 116
+RG + G ++ PVK+V NPDGSLA+AA L+KER++ + QAQ +D+ K S
Sbjct 421 LRGYG-KGGAEIEPVKVVKNPDGSLAQAALMQGALSKERKETKIQAQRERDMDT-QKGFS 478
Query 117 RPWEDPNPQPGERTIAQAL---QGLGQTGYEMPEW 148
+P G ++ A + + EMPEW
Sbjct 479 SNARILDPMSGNQSTAWSADESKDRNNKMKEMPEW 513
> tpv:TP01_0544 RNA helicase
Length=910
Score = 63.9 bits (154), Expect = 2e-10, Method: Composition-based stats.
Identities = 36/101 (35%), Positives = 59/101 (58%), Gaps = 5/101 (4%)
Query 1 RKLMSDYDKWEAQQLQRSGLVSSKENPYYDEEAGGILPAQEAEEDVEIEVVDEEPLFIRG 60
R+ ++D ++WE QQL +SG++SS+E E E E E+ + P F++G
Sbjct 5 RRYINDIERWENQQLLKSGILSSEEKSRLKAEL-----ELEEEPQPEVTINLNPPNFLKG 59
Query 61 QTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQ 101
QT R+G+ LSP+K+VA P+GSL R T+ + E + + +
Sbjct 60 QTIRSGIVLSPIKLVAKPEGSLQRTITTSLQIQNELKSLNR 100
> mmu:27060 Tcirg1, ATP6N1C, ATP6a3, Atp6i, OC-116, OPTB1, Stv1,
TIRC7, Vph1, oc; T-cell, immune regulator 1, ATPase, H+ transporting,
lysosomal V0 protein A3 (EC:3.6.3.6); K02154 V-type
H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=834
Score = 35.0 bits (79), Expect = 0.090, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query 57 FIRGQTTRAGMQLSPVK--IVANPDGSLARAAATAQTLAKERRDVRQAQEA 105
F+R + RAG+ L+P + + A P L R LA+E RDVR Q+A
Sbjct 67 FLREEVQRAGLTLAPPEGTLPAPPPRDLLRIQEETDRLAQELRDVRGNQQA 117
> hsa:10312 TCIRG1, ATP6N1C, ATP6V0A3, Atp6i, OC-116kDa, OC116,
OPTB1, Stv1, TIRC7, Vph1, a3; T-cell, immune regulator 1,
ATPase, H+ transporting, lysosomal V0 subunit A3 (EC:3.6.3.6);
K02154 V-type H+-transporting ATPase subunit I [EC:3.6.3.14]
Length=830
Score = 33.9 bits (76), Expect = 0.21, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query 57 FIRGQTTRAGMQLSPVK--IVANPDGSLARAAATAQTLAKERRDVRQAQEA 105
F++ + RAG+ L P K + A P L R + LA+E RDVR Q+A
Sbjct 67 FLQEEVRRAGLVLPPPKGRLPAPPPRDLLRIQEETERLAQELRDVRGNQQA 117
> ath:AT5G13010 EMB3011 (embryo defective 3011); ATP binding /
RNA helicase/ helicase/ nucleic acid binding; K12815 pre-mRNA-splicing
factor ATP-dependent RNA helicase PRP16 [EC:3.6.4.13]
Length=1226
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 46/105 (43%), Gaps = 11/105 (10%)
Query 2 KLMSDYDKWEAQQLQRSGLVSSKE-NPYYDEEAGGILPAQEAEEDVEIEVVDEEPLFIRG 60
+L +D +WE +QL RSG V E +D E E + V D +P F+ G
Sbjct 367 QLNADNAQWEDRQLLRSGAVRGTEVQTEFD---------SEEERKAILLVHDTKPPFLDG 417
Query 61 QTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEA 105
+ Q PV V +P +A + L KE R+ + A ++
Sbjct 418 RVVYTK-QAEPVMPVKDPTSDMAIISRKGSGLVKEIREKQSANKS 461
> sce:YER013W PRP22; DEAH-box RNA-dependent ATPase/ATP-dependent
RNA helicase, associates with lariat intermediates before
the second catalytic step of splicing; mediates ATP-dependent
mRNA release from the spliceosome and unwinds RNA duplexes
(EC:3.6.1.-); K12818 ATP-dependent RNA helicase DHX8/PRP22
[EC:3.6.4.13]
Length=1145
Score = 29.6 bits (65), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 35/149 (23%), Positives = 61/149 (40%), Gaps = 29/149 (19%)
Query 1 RKLMSDYDKWEAQQLQRSGLVSSKENPYYDEEA-------------GGI----------- 36
R+ ++ ++WE +QL SG S + P +E G I
Sbjct 288 RRALTSPERWEIRQLIASGAASIDDYPELKDEIPINTSYLTAKRDDGSIVNGNTEKVDSK 347
Query 37 LPAQEAEEDVEIEV---VDEEPLFIRGQTTRAGMQLSPVKIVANPDGSLARAAATAQTLA 93
L Q+ +E EI+V D+ P F++ Q + + KI P G + R+A
Sbjct 348 LEEQQRDETDEIDVELNTDDGPKFLKDQQVKGAKKYEMPKITKVPRGFMNRSAINGSNAI 407
Query 94 KERRDVRQAQEAAILDSIPKDMSRPWEDP 122
++ R+ + ++ I I K S ++DP
Sbjct 408 RDHREEKLRKKREIEQQIRKQQS--FDDP 434
> cel:C32C4.1 hypothetical protein
Length=538
Score = 29.3 bits (64), Expect = 4.9, Method: Composition-based stats.
Identities = 15/59 (25%), Positives = 30/59 (50%), Gaps = 0/59 (0%)
Query 52 DEEPLFIRGQTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDS 110
D +P+ G+ + + ++A P + ATAQ A E + ++QAQ +A+ ++
Sbjct 457 DAKPVTTLGKLVATSTSICGIIVLAFPISMIVEKFATAQQRAIEDQQIQQAQMSAVANN 515
> tpv:TP01_0641 RNA helicase
Length=974
Score = 29.3 bits (64), Expect = 5.3, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 94 KERRDVRQAQEAAILDSIPKDMSRPWEDPNPQPGERTI 131
K R+D+R+ +I S+P DM +P P+ + I
Sbjct 543 KNRKDIRELIILSIYSSLPSDMQNKIFEPTPENSRKVI 580
> tgo:TGME49_006180 hypothetical protein
Length=148
Score = 28.9 bits (63), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 27/61 (44%), Gaps = 9/61 (14%)
Query 70 SPVKIVANPDGSLARAAATAQTLAKERRDVRQAQEAAILDSIPKDMSRPWEDPNPQPGER 129
SP NPD RAA T++T + A SIP+ RP P+P G++
Sbjct 89 SPSSWDTNPDHPHLRAAVTSKT---------SSNTDAAFTSIPRPHHRPLFSPSPPHGDK 139
Query 130 T 130
+
Sbjct 140 S 140
> mmu:270110 Irf2bp2, E130305N23Rik; interferon regulatory factor
2 binding protein 2
Length=570
Score = 28.9 bits (63), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 58 IRGQTTRAGMQLSPVKIVANPDGSLARAAATAQTLAKER 96
+ GQ T + L PV + PD SLA +A TL ER
Sbjct 457 VGGQATGSTGGLEPVHPASLPDSSLAASAPLCCTLCHER 495
> eco:b3482 rhsB, ECK3466, JW5679; RhsB element core protein RshB
Length=1411
Score = 28.9 bits (63), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 18/84 (21%), Positives = 38/84 (45%), Gaps = 20/84 (23%)
Query 4 MSDYDKWEAQQLQRSGLVSSKENPY------------YDEEAGGILPAQEAEEDVEIEVV 51
++YD+W G + ++ENP+ YDEE+G + ++ +
Sbjct 1167 CAEYDEW--------GNLLNEENPHQLQQLIRLPGQQYDEESGLYYNRHRYYDPLQGRYI 1218
Query 52 DEEPLFIRGQTTRAGMQLSPVKIV 75
++P+ ++G G QL+P+ +
Sbjct 1219 TQDPIGLKGGWNLYGYQLNPISDI 1242
Lambda K H
0.310 0.129 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40