bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
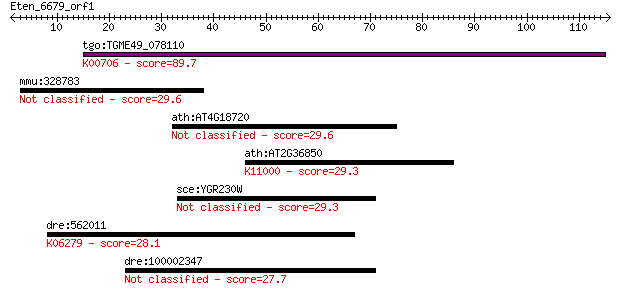
Query= Eten_6679_orf1
Length=115
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_078110 1,3-beta-glucan synthase component domain-co... 89.7 2e-18
mmu:328783 Mslnl, 4732467B22; mesothelin-like 29.6 2.2
ath:AT4G18720 transcription elongation factor-related 29.6 2.4
ath:AT2G36850 GSL8; GSL8 (GLUCAN SYNTHASE-LIKE 8); 1,3-beta-gl... 29.3 3.0
sce:YGR230W BNS1; Bns1p 29.3 3.3
dre:562011 shc2, MGC162768, si:dkey-94n12.3, zgc:162768; SHC (... 28.1 6.7
dre:100002347 hypothetical LOC100002347 27.7 8.5
> tgo:TGME49_078110 1,3-beta-glucan synthase component domain-containing
protein (EC:2.4.1.34); K00706 1,3-beta-glucan synthase
[EC:2.4.1.34]
Length=2321
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/103 (42%), Positives = 67/103 (65%), Gaps = 3/103 (2%)
Query 15 DARPKDEKPRRMG---LSVSRENLNAVIAELSCSSKPGSGVKTFVEHRTYAQVLRNFWRV 71
DA +++P G LS +RENLN + +L +KP G+KTF+E RTY QVLR+FWRV
Sbjct 277 DASIAEDRPGTSGGPRLSFTRENLNMFVHKLLNGTKPSEGIKTFMERRTYLQVLRSFWRV 336
Query 72 FAWHVTTFAGILLLYAVVDEESSYSVSKQWSILTLTAILSYGM 114
AWH TF+ + L AVVD+ES+ ++ W+ +T+++ + +
Sbjct 337 IAWHGVTFSLLFFLKAVVDDESTAELAFTWNRTVVTSVVLHAL 379
> mmu:328783 Mslnl, 4732467B22; mesothelin-like
Length=685
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query 3 DTRSFSDSFLRSDARPKDEKP-RRMGLSVSRENLNA 37
D R F D+FL S A +P RR G S R N+ A
Sbjct 283 DARRFVDNFLESKATSVSSRPKRRTGRSCVRGNITA 318
> ath:AT4G18720 transcription elongation factor-related
Length=266
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 11/48 (22%), Positives = 27/48 (56%), Gaps = 5/48 (10%)
Query 32 RENLNAVIAELSCSSKPGSGVKTFVEH-----RTYAQVLRNFWRVFAW 74
+E +++ ++ C + G G++ F++H R+ ++LR+ W F +
Sbjct 40 KEAPKSLVCDVVCKTSMGQGLEFFIDHKNPKIRSEGRILRDLWMKFFY 87
> ath:AT2G36850 GSL8; GSL8 (GLUCAN SYNTHASE-LIKE 8); 1,3-beta-glucan
synthase/ transferase, transferring glycosyl groups;
K11000 callose synthase [EC:2.4.1.-]
Length=1909
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 4/44 (9%)
Query 46 SKPGSGVKT----FVEHRTYAQVLRNFWRVFAWHVTTFAGILLL 85
SKP +T FVEHRTY + R+F R++ + F + ++
Sbjct 466 SKPKGRKRTAKSSFVEHRTYLHLFRSFIRLWIFMFIMFQSLTII 509
> sce:YGR230W BNS1; Bns1p
Length=137
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 33 ENLNAVIAELSCSSKPGSGVKTFVEHRTYAQVLRNFWR 70
ENL++ L S K KTF +H + QV +N +R
Sbjct 31 ENLDSFNQHLKMSLKVAHNTKTFAKHCLHRQVFKNTYR 68
> dre:562011 shc2, MGC162768, si:dkey-94n12.3, zgc:162768; SHC
(Src homology 2 domain containing) transforming protein 2;
K06279 src homology 2 domain-containing transforming protein
C
Length=501
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Query 8 SDSFLRSDARPKDEKPRRMGLSVSRENLNAVIAELSCSSKPGSGVKTFVEHRTYAQVLR 66
SD +LR+D P + L V+ +NL+ + A + +P S K + R + LR
Sbjct 313 SDGYLRADGHPPGSRDYEEHLYVNTQNLDNLEASV-VGRRPESPKKDIFDMRPFEDALR 370
> dre:100002347 hypothetical LOC100002347
Length=343
Score = 27.7 bits (60), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 23 PRRMGLSVSRENLNAVIAELSCSSKPGSGVKTFVEHRTYAQVLRNFWR 70
PR L + R + + + +KP S V F +RT +VLR W+
Sbjct 155 PRDRELGIERAHRALQLKPSNPEAKPRSKVVKFSSYRTKEEVLRKAWQ 202
Lambda K H
0.320 0.130 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2042676840
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40