bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6652_orf2
Length=118
Score E
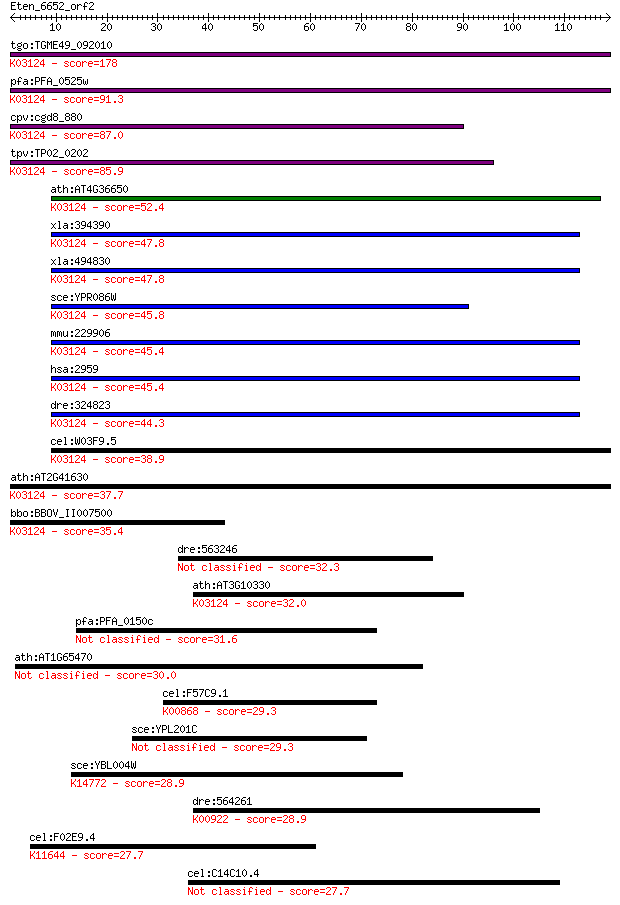
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_092010 transcription initiation factor IIB, putativ... 178 4e-45
pfa:PFA_0525w transcription initiation factor TFIIB, putative;... 91.3 8e-19
cpv:cgd8_880 transcription initiation factor TFIIB Sua7p; ZnR+... 87.0 1e-17
tpv:TP02_0202 transcription initiation factor TFIIB; K03124 tr... 85.9 3e-17
ath:AT4G36650 ATPBRP; ATPBRP (PLANT-SPECIFIC TFIIB-RELATED PRO... 52.4 3e-07
xla:394390 gtf2b-a, XLTFIIB, gtf2b, tf2b, tfiib; general trans... 47.8 9e-06
xla:494830 gtf2b-b, gtf2b, tf2b, tfiib; general transcription ... 47.8 1e-05
sce:YPR086W SUA7, SOH4; Sua7p; K03124 transcription initiation... 45.8 3e-05
mmu:229906 Gtf2b, MGC6859; general transcription factor IIB; K... 45.4 4e-05
hsa:2959 GTF2B, TF2B, TFIIB; general transcription factor IIB;... 45.4 4e-05
dre:324823 gtf2b, wu:fc44c11, zgc:66258; general transcription... 44.3 1e-04
cel:W03F9.5 ttb-1; TFII(two)B (general transcription factor) f... 38.9 0.004
ath:AT2G41630 TFIIB; TFIIB (TRANSCRIPTION FACTOR II B); RNA po... 37.7 0.010
bbo:BBOV_II007500 18.m06622; transcription initiation factor T... 35.4 0.045
dre:563246 si:dkey-45k15.2 32.3 0.33
ath:AT3G10330 transcription initiation factor IIB-2 / general ... 32.0 0.43
pfa:PFA_0150c conserved Plasmodium protein, unknown function 31.6 0.57
ath:AT1G65470 FAS1; FAS1 (FASCIATA 1); histone binding 30.0 1.9
cel:F57C9.1 hypothetical protein; K00868 pyridoxine kinase [EC... 29.3 3.0
sce:YPL201C YIG1; Protein that interacts with glycerol 3-phosp... 29.3 3.6
sce:YBL004W UTP20; Component of the small-subunit (SSU) proces... 28.9 4.7
dre:564261 pik3cg; phosphoinositide-3-kinase, catalytic, gamma... 28.9 4.7
cel:F02E9.4 sin-3; SIN3 (yeast Switch INdependent) histone dea... 27.7 8.9
cel:C14C10.4 hypothetical protein 27.7 9.8
> tgo:TGME49_092010 transcription initiation factor IIB, putative
; K03124 transcription initiation factor TFIIB
Length=464
Score = 178 bits (451), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 88/124 (70%), Positives = 104/124 (83%), Gaps = 10/124 (8%)
Query 1 ELVVYDRAVKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEH 60
ELVVYDRA+ EKELGK IN+IKKLLP RGG+N++ESA+ LLPRYCS+LQLS+HV+DVAEH
Sbjct 298 ELVVYDRAISEKELGKAINRIKKLLPQRGGVNSAESATQLLPRYCSRLQLSMHVADVAEH 357
Query 61 VAKRAAQVFVCSH------RPNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTGA 114
VAKRA QV + SH RPNSVAAAAIWLVV+LL+SS++ LPK +E+ASVTGA
Sbjct 358 VAKRATQVIISSHRHVLARRPNSVAAAAIWLVVKLLNSSTNPN----LPKASEIASVTGA 413
Query 115 GEHT 118
GEHT
Sbjct 414 GEHT 417
> pfa:PFA_0525w transcription initiation factor TFIIB, putative;
K03124 transcription initiation factor TFIIB
Length=367
Score = 91.3 bits (225), Expect = 8e-19, Method: Composition-based stats.
Identities = 46/118 (38%), Positives = 81/118 (68%), Gaps = 4/118 (3%)
Query 1 ELVVYDRAVKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEH 60
EL+ +DR+ KEK+LGK INK+KK+LP+R + N E+ SHL+ ++LQLS+ + + E+
Sbjct 210 ELITFDRSYKEKDLGKTINKLKKVLPSRAFVYN-ENISHLIYSLSNRLQLSIDLIEAIEY 268
Query 61 VAKRAAQVFVCSHRPNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTGAGEHT 118
V K+A+ + SHR NS+ +I L+V+L +++ ++ + LP ++++A+V G +T
Sbjct 269 VVKKASTLITTSHRLNSLCGGSIHLIVEL---NTNEEKNMKLPNLSQIATVCGVTTNT 323
> cpv:cgd8_880 transcription initiation factor TFIIB Sua7p; ZnR+2cyclins
; K03124 transcription initiation factor TFIIB
Length=456
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 44/89 (49%), Positives = 60/89 (67%), Gaps = 1/89 (1%)
Query 1 ELVVYDRAVKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEH 60
EL+ +DR + E+EL + INK+KK LP RG +S SA+ L+PR+C LQLS + +AE+
Sbjct 310 ELLSFDRTISERELVRSINKLKKDLPRRGPTLSS-SAAELMPRFCHYLQLSHEIVGIAEY 368
Query 61 VAKRAAQVFVCSHRPNSVAAAAIWLVVQL 89
V + A Q SHRPNS+AA AI+ V L
Sbjct 369 VCRTAEQYINKSHRPNSLAAGAIYFVCNL 397
> tpv:TP02_0202 transcription initiation factor TFIIB; K03124
transcription initiation factor TFIIB
Length=342
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 40/95 (42%), Positives = 61/95 (64%), Gaps = 1/95 (1%)
Query 1 ELVVYDRAVKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEH 60
ELV+YDR++ KELG+ IN++KK+LP RG +E S L+PR+CS+L LS + E
Sbjct 176 ELVIYDRSLTVKELGRAINRLKKVLPNRGNAP-TEDVSQLMPRFCSRLNLSNEIMTTCEA 234
Query 61 VAKRAAQVFVCSHRPNSVAAAAIWLVVQLLSSSSS 95
VA+++ + +HR S+A I+LV +L + S
Sbjct 235 VAQKSFVLLTSAHRTTSLAGGIIYLVTRLFFADDS 269
> ath:AT4G36650 ATPBRP; ATPBRP (PLANT-SPECIFIC TFIIB-RELATED PROTEIN);
RNA polymerase II transcription factor/ rDNA binding;
K03124 transcription initiation factor TFIIB
Length=369
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 57/109 (52%), Gaps = 11/109 (10%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQV 68
V++KE+GK I + + L IN++ + H+ PR+C+ LQL+ ++A H+ +
Sbjct 202 VQQKEIGKYIKILGEALQLSQPINSNSISVHM-PRFCTLLQLNKSAQELATHIGEVVINK 260
Query 69 FVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTGAGE 116
C+ R P S++AAAI+L QL + AE+ +TG E
Sbjct 261 CFCTRRNPISISAAAIYLACQLEDKRKTQ---------AEICKITGLTE 300
> xla:394390 gtf2b-a, XLTFIIB, gtf2b, tf2b, tfiib; general transcription
factor 2B; K03124 transcription initiation factor
TFIIB
Length=316
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 56/105 (53%), Gaps = 7/105 (6%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQV 68
+ +KE+G+ I K L T + + + R+CS L L+ V A H+A++A ++
Sbjct 186 ISKKEIGRCFKLILKALETNVDL---ITTGDFMSRFCSNLGLTKQVQMAATHIARKAVEL 242
Query 69 FVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVT 112
+ R P SVAAAAI++ Q +S+ + Q + +A VA VT
Sbjct 243 DLVPGRSPISVAAAAIYMASQ---ASAEKRTQKEIGDIAGVADVT 284
> xla:494830 gtf2b-b, gtf2b, tf2b, tfiib; general transcription
factor 2B; K03124 transcription initiation factor TFIIB
Length=316
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 56/105 (53%), Gaps = 7/105 (6%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQV 68
+ +KE+G+ I K L T + + + R+CS L L+ V A H+A++A ++
Sbjct 186 ISKKEIGRCFKLILKALETNVDL---ITTGDFMSRFCSNLGLTKQVQMAATHIARKAVEL 242
Query 69 FVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVT 112
+ R P SVAAAAI++ Q +S+ + Q + +A VA VT
Sbjct 243 DLVPGRSPISVAAAAIYMASQ---ASAEKRTQKEIGDIAGVADVT 284
> sce:YPR086W SUA7, SOH4; Sua7p; K03124 transcription initiation
factor TFIIB
Length=345
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 49/94 (52%), Gaps = 14/94 (14%)
Query 9 VKEKELGKQINKIKKLLPTRG---------GINNSESASHL--LPRYCSKLQLSVHVSDV 57
VK KE GK +N +K +L RG +N A +L +PR+CS L L + V+
Sbjct 198 VKTKEFGKTLNIMKNIL--RGKSEDGFLKIDTDNMSGAQNLTYIPRFCSHLGLPMQVTTS 255
Query 58 AEHVAKRAAQVF-VCSHRPNSVAAAAIWLVVQLL 90
AE+ AK+ ++ + P ++A +I+L + L
Sbjct 256 AEYTAKKCKEIKEIAGKSPITIAVVSIYLNILLF 289
> mmu:229906 Gtf2b, MGC6859; general transcription factor IIB;
K03124 transcription initiation factor TFIIB
Length=316
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 55/105 (52%), Gaps = 7/105 (6%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQV 68
+ +KE+G+ I K L T + + + R+CS L L V A H+A++A ++
Sbjct 186 ISKKEIGRCFKLILKALETSVDL---ITTGDFMSRFCSNLCLPKQVQMAATHIARKAVEL 242
Query 69 FVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVT 112
+ R P SVAAAAI++ Q +S+ + Q + +A VA VT
Sbjct 243 DLVPGRSPISVAAAAIYMASQ---ASAEKRTQKEIGDIAGVADVT 284
> hsa:2959 GTF2B, TF2B, TFIIB; general transcription factor IIB;
K03124 transcription initiation factor TFIIB
Length=316
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 55/105 (52%), Gaps = 7/105 (6%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQV 68
+ +KE+G+ I K L T + + + R+CS L L V A H+A++A ++
Sbjct 186 ISKKEIGRCFKLILKALETSVDL---ITTGDFMSRFCSNLCLPKQVQMAATHIARKAVEL 242
Query 69 FVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVT 112
+ R P SVAAAAI++ Q +S+ + Q + +A VA VT
Sbjct 243 DLVPGRSPISVAAAAIYMASQ---ASAEKRTQKEIGDIAGVADVT 284
> dre:324823 gtf2b, wu:fc44c11, zgc:66258; general transcription
factor IIB; K03124 transcription initiation factor TFIIB
Length=316
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 55/105 (52%), Gaps = 7/105 (6%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQV 68
+ +KE+G+ I K L T + + + R+CS L L V A ++A++A ++
Sbjct 186 ISKKEIGRCFKLILKALETSVDL---ITTGDFMSRFCSNLGLPKQVQMAATYIARKAVEL 242
Query 69 FVCSHR-PNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVT 112
+ R P SVAAAAI++ Q +S+ + Q + +A VA VT
Sbjct 243 DLVPGRSPISVAAAAIYMASQ---ASAEKKTQKEIGDIAGVADVT 284
> cel:W03F9.5 ttb-1; TFII(two)B (general transcription factor)
family member (ttb-1); K03124 transcription initiation factor
TFIIB
Length=306
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 52/113 (46%), Gaps = 17/113 (15%)
Query 9 VKEKELGKQINKIKKLLPTRGGINNSE--SASHLLPRYCSKLQLSVHVSDVAEHVAKRAA 66
V +KE+G+ I + L T N E +++ + R+C L L + A +AK A
Sbjct 176 VSKKEIGRCFKIIVRSLET-----NLEQITSADFMSRFCGNLSLPNSIQAAATRIAKCAV 230
Query 67 QV-FVCSHRPNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTGAGEHT 118
+ V P S+AAAAI++ Q + S++ E+ V GA E T
Sbjct 231 DMDLVAGRTPISIAAAAIYMASQASAEKRSAK---------EIGDVAGAAEIT 274
> ath:AT2G41630 TFIIB; TFIIB (TRANSCRIPTION FACTOR II B); RNA
polymerase II transcription factor/ protein binding / transcription
regulator/ translation initiation factor/ zinc ion binding;
K03124 transcription initiation factor TFIIB
Length=312
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 31/121 (25%), Positives = 55/121 (45%), Gaps = 13/121 (10%)
Query 1 ELVVYDRAVKEKELGKQINKIKKLLPTRGGIN---NSESASHLLPRYCSKLQLSVHVSDV 57
E+ V +KE+G+ + I K L G + + A + R+CS L +S H
Sbjct 170 EICVIANGATKKEIGRAKDYIVKTLGLEPGQSVDLGTIHAGDFMRRFCSNLAMSNHAVKA 229
Query 58 AEHVAKRAAQVFVCSHRPNSVAAAAIWLVVQLLSSSSSSQQQLALPKVAEVASVTGAGEH 117
A+ +++ + F P S+AA I+++ QL S ++ L +++ TG E
Sbjct 230 AQEAVQKSEE-FDIRRSPISIAAVVIYIITQL----SDDKKTL-----KDISHATGVAEG 279
Query 118 T 118
T
Sbjct 280 T 280
> bbo:BBOV_II007500 18.m06622; transcription initiation factor
TFIIB; K03124 transcription initiation factor TFIIB
Length=253
Score = 35.4 bits (80), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 1 ELVVYDRAVKEKELGKQINKIKKLLPTRGGINNSESASHLLP 42
EL +Y+ + +++L + I ++KKLLP RG +SA ++P
Sbjct 178 ELTIYEAKISQRDLSRAIGRMKKLLPQRGNATVEDSA-QIIP 218
> dre:563246 si:dkey-45k15.2
Length=787
Score = 32.3 bits (72), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 34 SESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFVCSHRPNSVAAAAI 83
SE + ++ P+Y VH V E+V++ + C +R N AAA I
Sbjct 613 SEQSKYMSPKYRQAFVNEVHFFVVKEYVSQLMKNNYSCKNRKNETAAAKI 662
> ath:AT3G10330 transcription initiation factor IIB-2 / general
transcription factor TFIIB-2 (TFIIB2); K03124 transcription
initiation factor TFIIB
Length=312
Score = 32.0 bits (71), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 37 ASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFVCSHRPNSVAAAAIWLVVQL 89
A + R+CS L ++ A+ +++ + F P S+AAA I+++ QL
Sbjct 209 AGDFMRRFCSNLGMTNQTVKAAQESVQKSEE-FDIRRSPISIAAAVIYIITQL 260
> pfa:PFA_0150c conserved Plasmodium protein, unknown function
Length=259
Score = 31.6 bits (70), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 32/59 (54%), Gaps = 5/59 (8%)
Query 14 LGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFVCS 72
L K +NK +++ P+ G ++ ++L +C KL L+ +D E + K+ Q+F S
Sbjct 67 LIKCLNKAEEMRPSFGSLD-----YNVLSNFCEKLFLAADKNDRNEVITKKTLQMFFTS 120
> ath:AT1G65470 FAS1; FAS1 (FASCIATA 1); histone binding
Length=815
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 41/83 (49%), Gaps = 5/83 (6%)
Query 2 LVVYDRAVKEKELGKQINKIKKLLPTRGGINNSESA-SHLLPRYCSKLQLSVHVSDVAEH 60
+V D + E E G Q++++ + P+ N + S P +C+ LQ H+ ++ +H
Sbjct 563 FMVPDGYLSEDE-GVQVDRMD-IDPSEQDANTTSSKQDQESPEFCALLQQQKHLQNLTDH 620
Query 61 VAKRAAQVFVC--SHRPNSVAAA 81
K+ + +C +H S+ AA
Sbjct 621 ALKKTQPLIICNLTHEKVSLLAA 643
> cel:F57C9.1 hypothetical protein; K00868 pyridoxine kinase [EC:2.7.1.35]
Length=348
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 22/43 (51%), Gaps = 1/43 (2%)
Query 31 INNSESASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQ-VFVCS 72
+NN + +H+L YC + ++DV + + K+ FVC
Sbjct 119 LNNINNYTHVLTGYCGNVTFLQKIADVVKDLKKKNGNTTFVCD 161
> sce:YPL201C YIG1; Protein that interacts with glycerol 3-phosphatase
and plays a role in anaerobic glycerol production;
localizes to the nucleus and cytosol
Length=461
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 23/51 (45%), Gaps = 5/51 (9%)
Query 25 LPTRGGINNSESA-----SHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFV 70
+PTR I NS S+ S + P + +L VH S +A +F+
Sbjct 341 IPTRDPIENSNSSPPVSDSEVYPIFYKTQELHVHASGTGRQIANNGKYIFI 391
> sce:YBL004W UTP20; Component of the small-subunit (SSU) processome,
which is involved in the biogenesis of the 18S rRNA;
K14772 U3 small nucleolar RNA-associated protein 20
Length=2493
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 30/66 (45%), Gaps = 4/66 (6%)
Query 13 ELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDV-AEHVAKRAAQVFVC 71
E G ++ +K LL R + N S LL RY L H SD +E + K Q+F
Sbjct 1765 EFGTLLSPVKALLMVRINLRNQNKLSELLRRYLLGLN---HNSDSESESILKFCHQLFQE 1821
Query 72 SHRPNS 77
S NS
Sbjct 1822 SEMSNS 1827
> dre:564261 pik3cg; phosphoinositide-3-kinase, catalytic, gamma
polypeptide; K00922 phosphatidylinositol-4,5-bisphosphate
3-kinase [EC:2.7.1.153]
Length=1069
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 32/68 (47%), Gaps = 12/68 (17%)
Query 37 ASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFVCSHRPNSVAAAAIWLVVQLLSSSSSS 96
+++L R+C K L +H+S VA S P+ + W +V L+ SSS
Sbjct 268 SNYLWVRHCLKNSLELHLSVVA------------ISSLPDDTVSTEYWPLVDSLTGLSSS 315
Query 97 QQQLALPK 104
++L L K
Sbjct 316 HEELCLKK 323
> cel:F02E9.4 sin-3; SIN3 (yeast Switch INdependent) histone deacetylase
component homolog family member (sin-3); K11644 paired
amphipathic helix protein Sin3a
Length=1507
Score = 27.7 bits (60), Expect = 8.9, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 33/57 (57%), Gaps = 4/57 (7%)
Query 5 YD-RAVKEKELGKQINKIKKLLPTRGGINNSESASHLLPRYCSKLQLSVHVSDVAEH 60
YD +A K K L +N+I+++ R N+++++ HL+ Y + ++ V+DV H
Sbjct 989 YDQKAFKSKPL---VNQIEQICEERRKNNSTDTSPHLILEYTPERKVYRDVNDVTGH 1042
> cel:C14C10.4 hypothetical protein
Length=1058
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Query 36 SASHLLPRYCSKLQLSVHVSDVAEHVAKRAAQVFVCSHRPNSVAAAAIWLVVQLLSSSSS 95
SA+ L C+K Q+ + V DV +HV + + PN + L L+ ++S
Sbjct 424 SANEFLITVCAKNQVPLFV-DVFDHVQQAGDIRNIFGQNPNLKRSIGRQLAQHLIKTASP 482
Query 96 SQQQLALPKVAEV 108
S++ + KVA V
Sbjct 483 SEKAALIAKVASV 495
Lambda K H
0.313 0.125 0.341
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40