bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6644_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
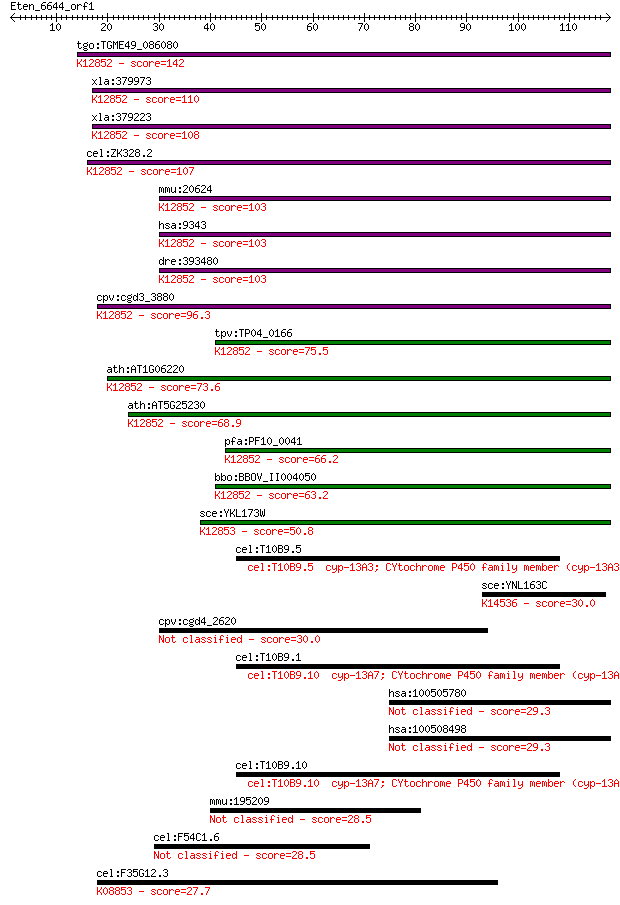
tgo:TGME49_086080 U5 small nuclear ribonucleoprotein, putative... 142 4e-34
xla:379973 snrp116-pending, MGC52678; U5 snRNP-specific protei... 110 2e-24
xla:379223 eftud2, MGC53479, snrp116, snu114; elongation facto... 108 3e-24
cel:ZK328.2 eft-1; Elongation FacTor family member (eft-1); K1... 107 8e-24
mmu:20624 Eftud2, 116kDa, Snrp116, U5-116kD; elongation factor... 103 1e-22
hsa:9343 EFTUD2, DKFZp686E24196, FLJ44695, KIAA0031, Snrp116, ... 103 1e-22
dre:393480 eftud2, MGC66214, wu:fi20f05, zgc:66214; elongation... 103 2e-22
cpv:cgd3_3880 Snu114p GTpase, U5 snRNP-specific protein, 116 k... 96.3 2e-20
tpv:TP04_0166 U5 small nuclear ribonucleoprotein; K12852 116 k... 75.5 4e-14
ath:AT1G06220 MEE5; MEE5 (MATERNAL EFFECT EMBRYO ARREST 5); GT... 73.6 2e-13
ath:AT5G25230 elongation factor Tu family protein; K12852 116 ... 68.9 4e-12
pfa:PF10_0041 U5 small nuclear ribonuclear protein, putative; ... 66.2 2e-11
bbo:BBOV_II004050 18.m06335; u5 small nuclear ribonuclear prot... 63.2 2e-10
sce:YKL173W SNU114, GIN10; GTPase component of U5 snRNP involv... 50.8 9e-07
cel:T10B9.5 cyp-13A3; CYtochrome P450 family member (cyp-13A3)... 30.4 1.5
sce:YNL163C RIA1, EFL1; Cytoplasmic GTPase involved in biogene... 30.0 1.7
cpv:cgd4_2620 hypothetical protein 30.0 1.8
cel:T10B9.1 cyp-13A4; CYtochrome P450 family member (cyp-13A4)... 29.6 2.8
hsa:100505780 hypothetical protein LOC100505780 29.3 2.9
hsa:100508498 hypothetical protein LOC100508498 29.3 2.9
cel:T10B9.10 cyp-13A7; CYtochrome P450 family member (cyp-13A7... 29.3 3.1
mmu:195209 Gm22; predicted gene 22 28.5
cel:F54C1.6 hypothetical protein 28.5 5.6
cel:F35G12.3 sel-5; Suppressor/Enhancer of Lin-12 family membe... 27.7 8.5
> tgo:TGME49_086080 U5 small nuclear ribonucleoprotein, putative
(EC:2.7.7.4); K12852 116 kDa U5 small nuclear ribonucleoprotein
component
Length=1008
Score = 142 bits (357), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 69/104 (66%), Positives = 82/104 (78%), Gaps = 2/104 (1%)
Query 14 LVRMDEDDSQQQQQKPVPHELKQYYPTAAEVYPEAETLVQEEDLQPITEPIIAPTKVSDF 73
LV + E++S VPHELK+YYP AEVYPEA+T+VQEED QPIT+PIIAP +DF
Sbjct 51 LVSIGEEESTAAAV--VPHELKKYYPDHAEVYPEADTVVQEEDTQPITQPIIAPVSTADF 108
Query 74 DLLETQLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
DLLE QLP T+FSFDYLASLM P +R++CLLG LHSGKTTF+
Sbjct 109 DLLEKQLPVTSFSFDYLASLMFQPESIRSVCLLGHLHSGKTTFL 152
> xla:379973 snrp116-pending, MGC52678; U5 snRNP-specific protein,
116 kD; K12852 116 kDa U5 small nuclear ribonucleoprotein
component
Length=974
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 53/102 (51%), Positives = 67/102 (65%), Gaps = 1/102 (0%)
Query 17 MDEDDSQQQQQKPVPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTKVSDFDL 75
M + D ++ + V HE K+YYPTA E+Y PE ET+VQEED QP+TEPII P K F +
Sbjct 47 MGDPDDERGGMEVVLHEDKKYYPTAEEIYGPEVETIVQEEDTQPLTEPIIKPVKAKKFSM 106
Query 76 LETQLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
+E LP T + D+LA LM P +RN+ L G LH GKT FV
Sbjct 107 MEQGLPATVYEMDFLADLMDNPELIRNVTLCGHLHHGKTCFV 148
> xla:379223 eftud2, MGC53479, snrp116, snu114; elongation factor
Tu GTP binding domain containing 2; K12852 116 kDa U5 small
nuclear ribonucleoprotein component
Length=974
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 53/102 (51%), Positives = 66/102 (64%), Gaps = 1/102 (0%)
Query 17 MDEDDSQQQQQKPVPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTKVSDFDL 75
M + D + + V HE K+YYPTA E+Y PE ET+VQEED QP+TEPII P K F +
Sbjct 47 MGDPDEDRGGMEVVLHEDKKYYPTAEEIYGPEVETIVQEEDTQPLTEPIIKPVKAKKFSV 106
Query 76 LETQLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
+E LP T + D+LA LM P +RN+ L G LH GKT FV
Sbjct 107 MEQALPATVYEMDFLADLMDNPELIRNVTLCGHLHHGKTCFV 148
> cel:ZK328.2 eft-1; Elongation FacTor family member (eft-1);
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=974
Score = 107 bits (268), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 56/104 (53%), Positives = 70/104 (67%), Gaps = 2/104 (1%)
Query 16 RMDEDDSQQ-QQQKPVPHELKQYYPTAAEVYPEA-ETLVQEEDLQPITEPIIAPTKVSDF 73
RM+EDD+++ Q + V HE K+YY TA EVY E ETLVQEED QP+TEPI+ P F
Sbjct 46 RMEEDDAEEIPQNQVVLHEDKKYYATALEVYGEGVETLVQEEDAQPLTEPIVKPVSKKKF 105
Query 74 DLLETQLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
E LPET + +YLA LM PH +RN+ + G LH GKTTF+
Sbjct 106 QAAERFLPETVYKKEYLADLMDCPHIMRNVAIAGHLHHGKTTFL 149
> mmu:20624 Eftud2, 116kDa, Snrp116, U5-116kD; elongation factor
Tu GTP binding domain containing 2; K12852 116 kDa U5 small
nuclear ribonucleoprotein component
Length=972
Score = 103 bits (257), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 52/89 (58%), Positives = 60/89 (67%), Gaps = 1/89 (1%)
Query 30 VPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFD 88
V HE K+YYPTA EVY PE ET+VQEED QP+TEPII P K F L+E LP T + D
Sbjct 58 VLHEDKKYYPTAEEVYGPEVETIVQEEDTQPLTEPIIKPVKTKKFTLMEQTLPVTVYEMD 117
Query 89 YLASLMHTPHCVRNICLLGSLHSGKTTFV 117
+LA LM +RN+ L G LH GKT FV
Sbjct 118 FLADLMDNSELIRNVTLCGHLHHGKTCFV 146
> hsa:9343 EFTUD2, DKFZp686E24196, FLJ44695, KIAA0031, Snrp116,
Snu114, U5-116KD; elongation factor Tu GTP binding domain
containing 2; K12852 116 kDa U5 small nuclear ribonucleoprotein
component
Length=937
Score = 103 bits (257), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 52/89 (58%), Positives = 60/89 (67%), Gaps = 1/89 (1%)
Query 30 VPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFD 88
V HE K+YYPTA EVY PE ET+VQEED QP+TEPII P K F L+E LP T + D
Sbjct 23 VLHEDKKYYPTAEEVYGPEVETIVQEEDTQPLTEPIIKPVKTKKFTLMEQTLPVTVYEMD 82
Query 89 YLASLMHTPHCVRNICLLGSLHSGKTTFV 117
+LA LM +RN+ L G LH GKT FV
Sbjct 83 FLADLMDNSELIRNVTLCGHLHHGKTCFV 111
> dre:393480 eftud2, MGC66214, wu:fi20f05, zgc:66214; elongation
factor Tu GTP binding domain containing 2; K12852 116 kDa
U5 small nuclear ribonucleoprotein component
Length=973
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 50/89 (56%), Positives = 63/89 (70%), Gaps = 1/89 (1%)
Query 30 VPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFD 88
V HE K+YYPTA EVY PE ET+VQEED QP+TEPII P ++ F L+E +LP T + +
Sbjct 59 VLHEDKKYYPTAEEVYGPEVETIVQEEDTQPLTEPIIKPVRMKQFTLMEQELPATVYDME 118
Query 89 YLASLMHTPHCVRNICLLGSLHSGKTTFV 117
+LA LM + +RN+ L G LH GKT FV
Sbjct 119 FLADLMDSSELIRNVTLCGHLHHGKTCFV 147
> cpv:cgd3_3880 Snu114p GTpase, U5 snRNP-specific protein, 116
kDa ; K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=1035
Score = 96.3 bits (238), Expect = 2e-20, Method: Composition-based stats.
Identities = 53/107 (49%), Positives = 68/107 (63%), Gaps = 7/107 (6%)
Query 18 DEDDSQQQQ----QKPVPHELKQYYPTAAEVY-PEAETLVQEEDLQPITEPIIAPTKVSD 72
DED Q+ K V E K+YYP ++VY + E LVQEED Q I EP+I+P K +
Sbjct 86 DEDHEMTQETDDDNKIVQFEDKEYYPDPSDVYGDDVEILVQEEDHQHIDEPLISPLKENK 145
Query 73 FDLLETQLPE--TTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
FDL+E L E TTFS+++L LM VRNIC +G +HSGKTTF+
Sbjct 146 FDLIEKNLKEMETTFSYEFLRDLMDNLEFVRNICFIGEIHSGKTTFL 192
> tpv:TP04_0166 U5 small nuclear ribonucleoprotein; K12852 116
kDa U5 small nuclear ribonucleoprotein component
Length=1028
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 38/94 (40%), Positives = 49/94 (52%), Gaps = 17/94 (18%)
Query 41 AAEVYPEAETLVQEEDLQPITEPIIAP-----------------TKVSDFDLLETQLPET 83
A + YP+ E V+EED Q I PII P V +FD+LE LP+
Sbjct 63 ADDEYPDVEVFVREEDTQTIEVPIIEPHEETVERVTNIKRLDSDVTVKNFDILEENLPQN 122
Query 84 TFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
FSF +L+SL P +RNIC+ G H GKTT +
Sbjct 123 KFSFQFLSSLTRKPEFIRNICICGGFHDGKTTLI 156
> ath:AT1G06220 MEE5; MEE5 (MATERNAL EFFECT EMBRYO ARREST 5);
GTP binding / GTPase/ translation elongation factor/ translation
factor, nucleic acid binding; K12852 116 kDa U5 small nuclear
ribonucleoprotein component
Length=987
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 42/99 (42%), Positives = 59/99 (59%), Gaps = 3/99 (3%)
Query 20 DDSQQQQQKPVPHELKQYYPTAAEVYPE-AETLVQEEDLQPITEPIIAPTKVSDFDLLET 78
+D + + Q +P + K+YYPTA EVY E ETLV +ED QP+ +PII P + F+ +
Sbjct 59 NDVEMENQIVLPED-KKYYPTAEEVYGEDVETLVMDEDEQPLEQPIIKPVRDIRFE-VGV 116
Query 79 QLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
+ T S +L LM P VRN+ L+G L GKT F+
Sbjct 117 KDQATYVSTQFLIGLMSNPALVRNVALVGHLQHGKTVFM 155
> ath:AT5G25230 elongation factor Tu family protein; K12852 116
kDa U5 small nuclear ribonucleoprotein component
Length=973
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 41/96 (42%), Positives = 55/96 (57%), Gaps = 2/96 (2%)
Query 24 QQQQKPVPHELKQYYPTAAEVYPE-AETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPE 82
+ Q V E K+YYP A EVY E ETLV +ED Q + +PII P + F++ +
Sbjct 46 NENQNIVLPEDKKYYPIAKEVYGEDVETLVMDEDEQSLEQPIIKPVRDIRFEVGVIKDQT 105
Query 83 TTF-SFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
TT+ S +L LM P VRN+ L+G L GKT F+
Sbjct 106 TTYVSTLFLIGLMSNPALVRNVALVGHLQHGKTVFM 141
> pfa:PF10_0041 U5 small nuclear ribonuclear protein, putative;
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=1235
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 36/92 (39%), Positives = 44/92 (47%), Gaps = 17/92 (18%)
Query 43 EVYPEAETLVQEEDLQPITEPIIAPTKVS-----------------DFDLLETQLPETTF 85
+VY E V+EED Q I E I + +FDL+ET LP F
Sbjct 91 KVYDGVEVFVEEEDTQDIEEATINKINANVERISFIKKLDVEANRKNFDLVETSLPNNNF 150
Query 86 SFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
SF YL+ LM +RNIC+ G H GKTT V
Sbjct 151 SFKYLSELMKQTSFIRNICIAGHFHHGKTTIV 182
> bbo:BBOV_II004050 18.m06335; u5 small nuclear ribonuclear protein;
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=999
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 48/94 (51%), Gaps = 17/94 (18%)
Query 41 AAEVYPEAETLVQEEDLQPITEPIIAP-----TKVSD------------FDLLETQLPET 83
A + + + E +Q ED Q I EPI+ +VS FD+LE ++P
Sbjct 57 ADDRFKDVEIFIQHEDTQTIDEPIVKSYETRVDRVSSIKRLDDDISAKCFDILEEKIPRN 116
Query 84 TFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
F+F ++ SLM P +RN+C+ G H GKTT +
Sbjct 117 RFTFHFMTSLMRQPQFIRNVCICGDFHHGKTTLI 150
> sce:YKL173W SNU114, GIN10; GTPase component of U5 snRNP involved
in mRNA splicing via spliceosome; binds directly to U5
snRNA; proposed to be involved in conformational changes of
the spliceosome; similarity to ribosomal translocation factor
EF-2; K12853 114 kDa U5 small nuclear ribonucleoprotein component
Length=1008
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 26/83 (31%), Positives = 46/83 (55%), Gaps = 3/83 (3%)
Query 38 YPTAAEVYPEAETLVQEEDLQPITEPIIAPTKVSD---FDLLETQLPETTFSFDYLASLM 94
+P EV ET + P+ EP+ TK+ + F L+ +P+T ++ DY+ S+
Sbjct 68 HPYGKEVEVLMETKNTQSPQTPLVEPVTERTKLQEHTIFTQLKKNIPKTRYNRDYMLSMA 127
Query 95 HTPHCVRNICLLGSLHSGKTTFV 117
+ P + N+ ++G LHSGKT+ +
Sbjct 128 NIPERIINVGVIGPLHSGKTSLM 150
> cel:T10B9.5 cyp-13A3; CYtochrome P450 family member (cyp-13A3);
K00517 [EC:1.14.-.-]
Length=520
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 34/63 (53%), Gaps = 1/63 (1%)
Query 45 YPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFDYLASLMHTPHCVRNIC 104
+PE + +QEE + +P + ++S +E + ET + LAS++H C+++
Sbjct 345 HPEIQKKLQEEVDRECPDPEVTFDQLSKLKYMECVIKETLRLYP-LASIVHNRKCMKSTT 403
Query 105 LLG 107
+LG
Sbjct 404 VLG 406
> sce:YNL163C RIA1, EFL1; Cytoplasmic GTPase involved in biogenesis
of the 60S ribosome; has similarity to translation elongation
factor 2 (Eft1p and Eft2p); K14536 ribosome assembly
protein 1 [EC:3.6.5.-]
Length=1110
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 93 LMHTPHCVRNICLLGSLHSGKTTF 116
L + P C+RNIC++ + GKT+
Sbjct 12 LQNDPSCIRNICIVAHVDHGKTSL 35
> cpv:cgd4_2620 hypothetical protein
Length=324
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/70 (25%), Positives = 33/70 (47%), Gaps = 7/70 (10%)
Query 30 VPHELKQYY------PTAAEVYPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPET 83
V H+ K + + EVY + + + + + P++ ++ D L ET+ PE
Sbjct 189 VEHDFKHIFSNDFQEKSLQEVYSQVRSKIHDSAINNKGIPLMFE-EILDEKLFETKEPEE 247
Query 84 TFSFDYLASL 93
F+FD+L S
Sbjct 248 DFNFDFLDSF 257
> cel:T10B9.1 cyp-13A4; CYtochrome P450 family member (cyp-13A4);
K00517 [EC:1.14.-.-]
Length=520
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Query 45 YPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFDYLASLMHTPHCVRNIC 104
+PE + +QEE + +P + ++S +E + E + LASL+H C++
Sbjct 345 HPEIQKKLQEEVDRECPDPEVTFDQISKLKYMECVVKEALRMYP-LASLVHNRKCMKKTN 403
Query 105 LLG 107
+LG
Sbjct 404 VLG 406
> hsa:100505780 hypothetical protein LOC100505780
Length=263
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 75 LLETQLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
LL+ ++ S D + L +RNIC+L + GKTT
Sbjct 125 LLQPEIIMVLNSLDKMIQLQKNTANIRNICVLAHVDRGKTTLA 167
> hsa:100508498 hypothetical protein LOC100508498
Length=263
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 75 LLETQLPETTFSFDYLASLMHTPHCVRNICLLGSLHSGKTTFV 117
LL+ ++ S D + L +RNIC+L + GKTT
Sbjct 125 LLQPEIIMVLNSLDKMIQLQKNTANIRNICVLAHVDRGKTTLA 167
> cel:T10B9.10 cyp-13A7; CYtochrome P450 family member (cyp-13A7);
K00517 [EC:1.14.-.-]
Length=518
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Query 45 YPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQLPETTFSFDYLASLMHTPHCVRNIC 104
+PE + +QEE + +P + ++S LE + E + LASL+H C++
Sbjct 345 HPEIQKKLQEEVDRECPDPEVTFDQLSKLKYLECVVKEALRLYP-LASLVHNRKCLKTTN 403
Query 105 LLG 107
+LG
Sbjct 404 VLG 406
> mmu:195209 Gm22; predicted gene 22
Length=3765
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 40 TAAEVYPEAETLVQEEDLQPITEPIIAPTKVSDFDLLETQL 80
+ ++ PE L +E D +P + P V DF+ L TQL
Sbjct 2849 SVGDMSPEPPNLEREHDNRPPGNASLPPLYVKDFEALSTQL 2889
> cel:F54C1.6 hypothetical protein
Length=280
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 26/46 (56%), Gaps = 4/46 (8%)
Query 29 PVPHELKQYYPT---AAEVYPEAETLVQEED-LQPITEPIIAPTKV 70
PVP E+ +PT ++ P AE + D L+PI EPI+ PT V
Sbjct 225 PVPPEVHTPHPTHDVKSKDEPSAEVISNTADILEPIQEPIMDPTPV 270
> cel:F35G12.3 sel-5; Suppressor/Enhancer of Lin-12 family member
(sel-5); K08853 AP2-associated kinase [EC:2.7.11.1]
Length=1077
Score = 27.7 bits (60), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 34/78 (43%), Gaps = 4/78 (5%)
Query 18 DEDDSQQQQQKPVPHELKQYYPTAAEVYPEAETLVQEEDLQPITEPIIAPTKVSDFDLLE 77
DE D Q ++ P+ +Q + ++ P AE + P++ P+I PT + DL +
Sbjct 439 DETDGSQVRKVPIDFHHRQSFSGEEQLKPAAEA----DSAGPLSCPLIKPTDLGFTDLDK 494
Query 78 TQLPETTFSFDYLASLMH 95
LP D L H
Sbjct 495 PALPRDRAQTDGKRRLPH 512
Lambda K H
0.319 0.133 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2032807080
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40