bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6588_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
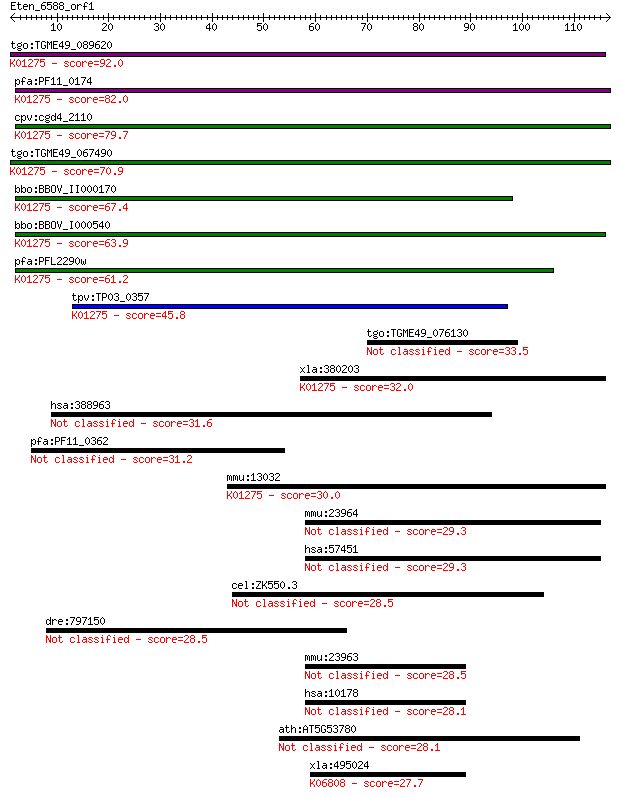
tgo:TGME49_089620 cathepsin C (EC:3.4.14.1); K01275 cathepsin ... 92.0 4e-19
pfa:PF11_0174 cathepsin C, homolog; K01275 cathepsin C [EC:3.4... 82.0 4e-16
cpv:cgd4_2110 preprocathepsin c precursor ; K01275 cathepsin C... 79.7 2e-15
tgo:TGME49_067490 papain family cysteine protease domain-conta... 70.9 1e-12
bbo:BBOV_II000170 18.m05995; cathepsin C precursor (EC:3.4.22.... 67.4 1e-11
bbo:BBOV_I000540 16.m00694; preprocathepsin c precursor; K0127... 63.9 1e-10
pfa:PFL2290w preprocathepsin c precursor, putative (EC:3.4.14.... 61.2 8e-10
tpv:TP03_0357 cathepsin C; K01275 cathepsin C [EC:3.4.14.1] 45.8 4e-05
tgo:TGME49_076130 cathepsin C2 (TgCPC2) (EC:3.4.14.1) 33.5 0.19
xla:380203 ctsc, MGC69126; cathepsin C (EC:3.4.14.1); K01275 c... 32.0 0.47
hsa:388963 C2orf81, hCG40743; chromosome 2 open reading frame 81 31.6
pfa:PF11_0362 protein phosphatase, putative 31.2 0.93
mmu:13032 Ctsc, AI047818, DPP1, DPPI; cathepsin C (EC:3.4.14.1... 30.0 1.8
mmu:23964 Odz2, 2610040L17Rik, 9330187F13Rik, D3Bwg1534e, Odz3... 29.3 2.8
hsa:57451 ODZ2, DKFZp686A1568, TEN-M2, TNM2; odz, odd Oz/ten-m... 29.3 2.8
cel:ZK550.3 hypothetical protein 28.5 5.8
dre:797150 hif1aa; hypoxia-inducible factor 1, alpha subunit (... 28.5 5.8
mmu:23963 Odz1, Odz2, TCAP-1, Ten-m1; odd Oz/ten-m homolog 1 (... 28.5 5.9
hsa:10178 ODZ1, ODZ3, TEN-M1, TNM, TNM1; odz, odd Oz/ten-m hom... 28.1 6.3
ath:AT5G53780 hypothetical protein 28.1 7.0
xla:495024 cdh13; cadherin 13, H-cadherin (heart) (EC:3.6.1.3)... 27.7 8.5
> tgo:TGME49_089620 cathepsin C (EC:3.4.14.1); K01275 cathepsin
C [EC:3.4.14.1]
Length=733
Score = 92.0 bits (227), Expect = 4e-19, Method: Composition-based stats.
Identities = 49/121 (40%), Positives = 71/121 (58%), Gaps = 9/121 (7%)
Query 1 APNKNTENLEAPVADSRTYLSRHGGVSTALELELSDETVPLNWVYDTK----DNLHRNEW 56
+PN NT NL + D +++LS GG+ L + L+D +P+++ +N HR W
Sbjct 65 SPNTNTSNLREELKDYKSFLSTQGGIKKELRVSLTD--IPVSFATTRTAAALENPHRRTW 122
Query 57 KMLAVKDKE--KKVVGGWTAVYDEGFEVHLTNKTRLFAILKYSVKQNCPEAKDGDLEDSN 114
K L V D + +K+VG WT VYDEGFEV + + RL +KY+ + NC DGDLE+S
Sbjct 123 KTLGVFDGQDKRKLVGSWTTVYDEGFEVDVGGRMRLMGFMKYNPQNNC-SIVDGDLENSQ 181
Query 115 G 115
G
Sbjct 182 G 182
> pfa:PF11_0174 cathepsin C, homolog; K01275 cathepsin C [EC:3.4.14.1]
Length=700
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 47/116 (40%), Positives = 70/116 (60%), Gaps = 4/116 (3%)
Query 2 PNKNTENLEAPVADSRTYLSRHGG-VSTALELELSDETVPLNWVYDTKDNLHRNEWKMLA 60
PNKNT N+ + D + YL + + L + LSD+ V +Y+T+DN HR++WK+LA
Sbjct 61 PNKNTYNV--GITDYKKYLLENNYEFVSELNVILSDDYVLYGDIYNTQDNEHRSKWKVLA 118
Query 61 VKDKEKKVVGGWTAVYDEGFEVHLTNKTRLFAILKYSVKQNCPEAKDGDLEDSNGE 116
V D+ K+V+G WT + DEGFE+ + N+T A++ Y C D D DSNG+
Sbjct 119 VYDENKRVIGTWTTICDEGFEIKIGNETYA-ALMHYEPNGKCGPVSDEDSLDSNGD 173
> cpv:cgd4_2110 preprocathepsin c precursor ; K01275 cathepsin
C [EC:3.4.14.1]
Length=635
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 54/121 (44%), Positives = 73/121 (60%), Gaps = 8/121 (6%)
Query 2 PNKNTENLEAPVADSRTYLS-RHGGVSTALELELSDETVPLNWVYDTKDNLH--RNEWKM 58
PNKNT+NL+ +AD +L+ + G L+L L+D N D ++ R++W
Sbjct 56 PNKNTQNLQPSLADYNKWLTTKTNGKLEKLDLILTDLLFSENDYSDLDRSMFPGRSDWTF 115
Query 59 LAVKDKEK-KVVGGWTAVYDEGFEVHLTNKTRLFAILKYSV--KQNCPEAKDGDLEDSNG 115
LAVKD E KVVG WT VYDEGFE+ + N T +F I+KY++ NC AKDGD E + G
Sbjct 116 LAVKDPENGKVVGRWTMVYDEGFEIVVNNMT-IFGIMKYNLLNNGNC-SAKDGDNETTKG 173
Query 116 E 116
E
Sbjct 174 E 174
> tgo:TGME49_067490 papain family cysteine protease domain-containing
protein (EC:3.4.14.1); K01275 cathepsin C [EC:3.4.14.1]
Length=622
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 55/144 (38%), Positives = 67/144 (46%), Gaps = 34/144 (23%)
Query 1 APNKNTENLEA------PVADSRTYLSRHGGVSTALELELSDETVPLNWVYDTKDNLHRN 54
APN N ENL +++ ++L+ V T +EL L+D+ + N D RN
Sbjct 42 APNFNLENLRPVYCVTPALSNYASWLASQSPVETEVELSLTDKLIRKN-----SDMAPRN 96
Query 55 EWKMLAVK--DKEKKVVGGWTAVYDEGFEVHLTNKTRLFAILKYSVK--QNCP------- 103
WK LAV K VG WT VYDEGFEV L N+ R F LKYS K + CP
Sbjct 97 NWKYLAVTLPSKPDVPVGTWTMVYDEGFEVRLPNR-RFFGFLKYSRKNGKECPRRLNGKS 155
Query 104 -----------EAKDGDLEDSNGE 116
A GD EDS GE
Sbjct 156 GVRSRKLRFKLRASVGDNEDSFGE 179
> bbo:BBOV_II000170 18.m05995; cathepsin C precursor (EC:3.4.22.-);
K01275 cathepsin C [EC:3.4.14.1]
Length=530
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 38/96 (39%), Positives = 55/96 (57%), Gaps = 5/96 (5%)
Query 2 PNKNTENLEAPVADSRTYLSRHGGVSTALELELSDETVPLNWVYDTKDNLHRNEWKMLAV 61
PN NL+ + + YL H + +LELS + +P + DT + +R +W+ LAV
Sbjct 67 PNTLEGNLK--LGNYLDYLKSHYTLDRNFQLELSLDVIPYS---DTSNKPNRYKWRALAV 121
Query 62 KDKEKKVVGGWTAVYDEGFEVHLTNKTRLFAILKYS 97
KD KVVG WT V D+GFEV L + TR F ++Y+
Sbjct 122 KDAAGKVVGRWTIVNDQGFEVLLDDSTRYFFYIRYT 157
> bbo:BBOV_I000540 16.m00694; preprocathepsin c precursor; K01275
cathepsin C [EC:3.4.14.1]
Length=546
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 41/118 (34%), Positives = 59/118 (50%), Gaps = 12/118 (10%)
Query 2 PNKNTENLEAPVADSRTYLSRHGGVSTALELELSDETVPLNWVYDTKDNLHRNEWKMLAV 61
PN+N +NL+ + D R YL H G +EL DE + + RN+W L V
Sbjct 29 PNRNVDNLK--LGDYRAYLENHYGKLNETVIELVDERY-----HRRSEAPPRNQWLYLGV 81
Query 62 KDKE--KKVVGGWTAVYDEGFEVHLTNKTRLFAILKYSV--KQNCPEAKDGDLEDSNG 115
++ + + V+G WT VYDEG ++ L T F KY+ +CP +G EDS G
Sbjct 82 RNPKEGQGVIGKWTMVYDEGLDIEL-GTTHYFGFFKYNKINSSSCPMIMEGSQEDSQG 138
> pfa:PFL2290w preprocathepsin c precursor, putative (EC:3.4.14.1);
K01275 cathepsin C [EC:3.4.14.1]
Length=590
Score = 61.2 bits (147), Expect = 8e-10, Method: Composition-based stats.
Identities = 37/107 (34%), Positives = 58/107 (54%), Gaps = 9/107 (8%)
Query 2 PNKNTENLEAPVADSRTYLSRHGGVSTALELELSDETVPLNWVYDTKDNLHRNEWKMLAV 61
PN+N +NL + + + +L G + + L+ E V + R++W LAV
Sbjct 54 PNRNLDNLNPSIRNYQRFLENEYGNLDMMIVNLTMEKVKI-----INQEKPRDKWTYLAV 108
Query 62 KDKEK-KVVGGWTAVYDEGFEVHLTNKTRLFAILKYSVKQN--CPEA 105
+D E+ +++G WT VYDEGFE+ L N ++ FA KY K N CP +
Sbjct 109 RDYERNEIIGHWTMVYDEGFEIRL-NGSKYFAFFKYERKSNAHCPTS 154
> tpv:TP03_0357 cathepsin C; K01275 cathepsin C [EC:3.4.14.1]
Length=501
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 45/84 (53%), Gaps = 4/84 (4%)
Query 13 VADSRTYLSRHGGVSTALELELSDETVPLNWVYDTKDNLHRNEWKMLAVKDKEKKVVGGW 72
+ D + ++ G+ T L+LEL+ + + V + K+ R+ W LAVK VVG W
Sbjct 38 LGDYKKLMTGDYGLGTTLDLELTLDKIDYKNVTNAKN---RDNWISLAVKS-HGYVVGHW 93
Query 73 TAVYDEGFEVHLTNKTRLFAILKY 96
T++YD+GF + L + L L +
Sbjct 94 TSIYDQGFRITLNDGLDLLVYLYF 117
> tgo:TGME49_076130 cathepsin C2 (TgCPC2) (EC:3.4.14.1)
Length=753
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 17/31 (54%), Gaps = 2/31 (6%)
Query 70 GGWTAVYDEG--FEVHLTNKTRLFAILKYSV 98
G WT VYDEG FEV R FA KY +
Sbjct 135 GLWTLVYDEGLHFEVSTPQNRRFFAFFKYQL 165
> xla:380203 ctsc, MGC69126; cathepsin C (EC:3.4.14.1); K01275
cathepsin C [EC:3.4.14.1]
Length=458
Score = 32.0 bits (71), Expect = 0.47, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 6/64 (9%)
Query 57 KMLAVKDKEKKVVGGWTAVYDEGFEVHLTNKTRLFAILKY-----SVKQNCPEAKDGDLE 111
K L + + + +G +T +Y++GFEV++ N LFA KY ++ C E G +
Sbjct 67 KELNIAEDQNGNLGSFTLIYNQGFEVNILNHL-LFAFFKYEQRGSNITSYCHETFPGWVH 125
Query 112 DSNG 115
D G
Sbjct 126 DVLG 129
> hsa:388963 C2orf81, hCG40743; chromosome 2 open reading frame
81
Length=588
Score = 31.6 bits (70), Expect = 0.58, Method: Composition-based stats.
Identities = 24/90 (26%), Positives = 40/90 (44%), Gaps = 5/90 (5%)
Query 9 LEAPVADSRTYLSRHGGVSTALELELSDETVPLNWVYDTKDNLHRNEWKMLAVKDKEKKV 68
+++P+ + R GV+ + ++ TVP+ V L EW L ++ + V
Sbjct 3 MQSPIVQRQERQVRDRGVTRSKAEKVRPPTVPVPQVDIVPGRLSEAEWMALTALEEGEDV 62
Query 69 VGGWTA-----VYDEGFEVHLTNKTRLFAI 93
VG A V D F+V+LT + F I
Sbjct 63 VGDILADLLARVMDSAFKVYLTQQCIPFTI 92
> pfa:PF11_0362 protein phosphatase, putative
Length=689
Score = 31.2 bits (69), Expect = 0.93, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 27/49 (55%), Gaps = 5/49 (10%)
Query 5 NTENLEAPVADSRTYLSRHGGVSTALELELSDETVPLNWVYDTKDNLHR 53
N N PV ++TY S HG +S ++S+ ++ LN Y KD LH+
Sbjct 297 NEHNNHNPVVRAKTYNSNHGVLS-----DMSNPSLGLNENYGFKDELHK 340
> mmu:13032 Ctsc, AI047818, DPP1, DPPI; cathepsin C (EC:3.4.14.1);
K01275 cathepsin C [EC:3.4.14.1]
Length=462
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 40/94 (42%), Gaps = 22/94 (23%)
Query 43 WVYDTKDNLHRNEWKMLAVKDKEKKVV----------------GGWTAVYDEGFEVHLTN 86
WV+ R++ ++ E+KVV G +T +Y++GFE+ L N
Sbjct 39 WVFQVGPRSSRSDINCSVMEATEEKVVVHLKKLDTAYDELGNSGHFTLIYNQGFEIVL-N 97
Query 87 KTRLFAILKYSVKQN-----CPEAKDGDLEDSNG 115
+ FA KY V+ + C E G + D G
Sbjct 98 DYKWFAFFKYEVRGHTAISYCHETMTGWVHDVLG 131
> mmu:23964 Odz2, 2610040L17Rik, 9330187F13Rik, D3Bwg1534e, Odz3,
Ten-m2, mKIAA1127; odd Oz/ten-m homolog 2 (Drosophila)
Length=2764
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Query 58 MLAVKDKEKKVVGGWTAVYDEGFEVHLTNKTRLFAILKYSVKQNCPEAKDGDLEDSN 114
+LA K E GWT YD E LTN TR ++ S+ + ++ D+E+SN
Sbjct 1678 LLATKSDET----GWTTFYDYDHEGRLTNVTRPTGVVT-SLHREMEKSITIDIENSN 1729
> hsa:57451 ODZ2, DKFZp686A1568, TEN-M2, TNM2; odz, odd Oz/ten-m
homolog 2 (Drosophila)
Length=2765
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Query 58 MLAVKDKEKKVVGGWTAVYDEGFEVHLTNKTRLFAILKYSVKQNCPEAKDGDLEDSN 114
+LA K E GWT YD E LTN TR ++ S+ + ++ D+E+SN
Sbjct 1679 LLATKSDET----GWTTFYDYDHEGRLTNVTRPTGVVT-SLHREMEKSITIDIENSN 1730
> cel:ZK550.3 hypothetical protein
Length=772
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 34/78 (43%), Gaps = 18/78 (23%)
Query 44 VYDTKDNLHR-----NEWKMLAVK--------DKEKKVVGGWTAVYDEGFEVHL-----T 85
+Y+ KD L EW +L +K DK +KV+G W+ DE ++ T
Sbjct 190 LYEAKDGLSEWQSRLLEWYLLEIKASGLDKHDDKTRKVLGSWSKFVDEYRSKYITGVMST 249
Query 86 NKTRLFAILKYSVKQNCP 103
N + F + V ++ P
Sbjct 250 NDQQTFVVTDQKVIKDAP 267
> dre:797150 hif1aa; hypoxia-inducible factor 1, alpha subunit
(basic helix-loop-helix transcription factor) a
Length=798
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 4/58 (6%)
Query 8 NLEAPVADSRTYLSRHGGVSTALELELSDETVPLNWVYDTKDNLHRNEWKMLAVKDKE 65
N+EAP+ DSRT+LSRH + + DE + +D +D L + ++ D +
Sbjct 310 NIEAPL-DSRTFLSRH---TLDMRFTYCDERITELLGFDPEDVLQHSVYEYYHALDSD 363
> mmu:23963 Odz1, Odz2, TCAP-1, Ten-m1; odd Oz/ten-m homolog 1
(Drosophila)
Length=2731
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 15/31 (48%), Positives = 17/31 (54%), Gaps = 4/31 (12%)
Query 58 MLAVKDKEKKVVGGWTAVYDEGFEVHLTNKT 88
+LA K E GWT VY+ E HLTN T
Sbjct 1645 LLATKSNEN----GWTTVYEYDPEGHLTNAT 1671
> hsa:10178 ODZ1, ODZ3, TEN-M1, TNM, TNM1; odz, odd Oz/ten-m homolog
1(Drosophila)
Length=2732
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 15/31 (48%), Positives = 17/31 (54%), Gaps = 4/31 (12%)
Query 58 MLAVKDKEKKVVGGWTAVYDEGFEVHLTNKT 88
+LA K E GWT VY+ E HLTN T
Sbjct 1646 LLATKSNEN----GWTTVYEYDPEGHLTNAT 1672
> ath:AT5G53780 hypothetical protein
Length=376
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 36/68 (52%), Gaps = 15/68 (22%)
Query 53 RNEWKMLAVKDKEKKVV---GG-------WTAVYDEGFEVHLTNKTRLFAILKYSVKQNC 102
++EW++LA+ K + +V GG + YD+G + + NKT+ F + K + K
Sbjct 241 QSEWELLAMCIKIEHLVESPGGRIFVVKQYVETYDQGDKEVIFNKTKRFMVFKLNTK--- 297
Query 103 PEAKDGDL 110
A DG++
Sbjct 298 --ATDGEI 303
> xla:495024 cdh13; cadherin 13, H-cadherin (heart) (EC:3.6.1.3);
K06808 cadherin 13, H-cadherin
Length=709
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 18/37 (48%), Gaps = 7/37 (18%)
Query 59 LAVKDKEKKVVGGWTAVYD-------EGFEVHLTNKT 88
L V+DK+ G W AVY + FE+H KT
Sbjct 379 LTVEDKDDPATGAWKAVYTIINGNPGQSFEIHTNPKT 415
Lambda K H
0.310 0.129 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2037741960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40