bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
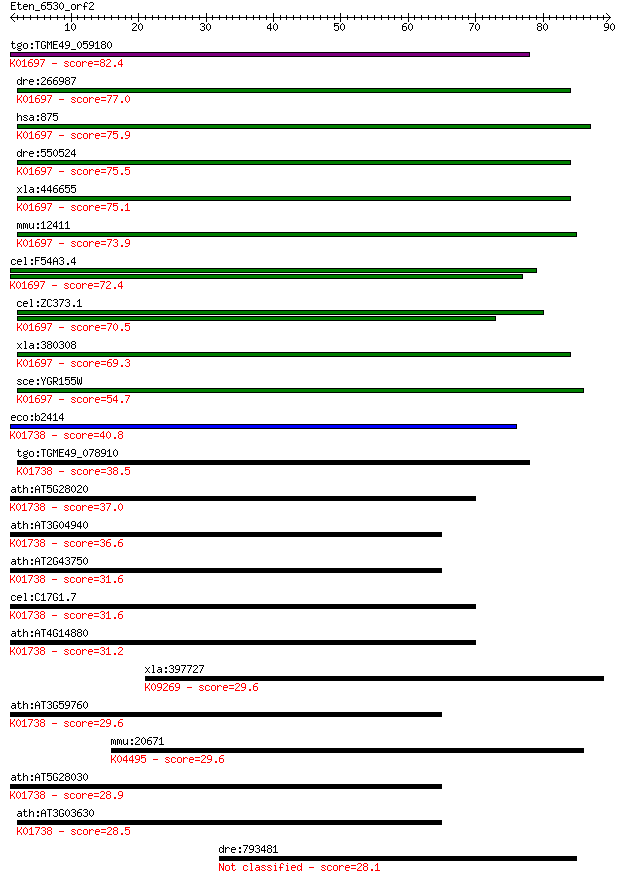
Query= Eten_6530_orf2
Length=89
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_059180 cystathionine beta-synthase, putative (EC:4.... 82.4 3e-16
dre:266987 cbsb, cb442, cbs, wu:fb37g05, wu:fm61c07, wu:fq06c0... 77.0 1e-14
hsa:875 CBS, HIP4; cystathionine-beta-synthase (EC:4.2.1.22); ... 75.9 3e-14
dre:550524 cbsa, MGC174292, zgc:110199; cystathionine-beta-syn... 75.5 3e-14
xla:446655 cbs-b, MGC82640, cbs, hip4; cystathionine-beta-synt... 75.1 5e-14
mmu:12411 Cbs, AI047524, AI303044, HIP4, MGC18856, MGC18895, M... 73.9 1e-13
cel:F54A3.4 hypothetical protein; K01697 cystathionine beta-sy... 72.4 3e-13
cel:ZC373.1 hypothetical protein; K01697 cystathionine beta-sy... 70.5 1e-12
xla:380308 cbs-a, MGC52737, hip4; cystathionine-beta-synthase ... 69.3 3e-12
sce:YGR155W CYS4, NHS5, STR4, VMA41; Cys4p (EC:4.2.1.22); K016... 54.7 8e-08
eco:b2414 cysK, cysZ, ECK2409, JW2407; cysteine synthase A, O-... 40.8 0.001
tgo:TGME49_078910 O-acetylserine (thiol) lyase, putative (EC:2... 38.5 0.005
ath:AT5G28020 CYSD2; CYSD2 (CYSTEINE SYNTHASE D2); catalytic/ ... 37.0 0.017
ath:AT3G04940 CYSD1; CYSD1 (CYSTEINE SYNTHASE D1); cysteine sy... 36.6 0.020
ath:AT2G43750 OASB; OASB (O-ACETYLSERINE (THIOL) LYASE B); cys... 31.6 0.69
cel:C17G1.7 hypothetical protein; K01738 cysteine synthase A [... 31.6 0.70
ath:AT4G14880 OASA1; OASA1 (O-ACETYLSERINE (THIOL) LYASE (OAS-... 31.2 0.77
xla:397727 sox13, Sry, sox12, xSox12, xsox13; SRY (sex determi... 29.6 2.2
ath:AT3G59760 OASC; OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM... 29.6 2.5
mmu:20671 Sox17, Sox; SRY-box containing gene 17; K04495 trans... 29.6 2.5
ath:AT5G28030 cysteine synthase, putative / O-acetylserine (th... 28.9 4.1
ath:AT3G03630 CS26; CS26; cysteine synthase; K01738 cysteine s... 28.5 5.7
dre:793481 sterile alpha motif domain containing 9-like 28.1 7.2
> tgo:TGME49_059180 cystathionine beta-synthase, putative (EC:4.2.1.22);
K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=514
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 41/77 (53%), Positives = 53/77 (68%), Gaps = 4/77 (5%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
VKV DAES +R +IR EG+ VGGSSGA + GAL+A ++ ++ ++LPD SR
Sbjct 262 VKVGDAESFTTARAIIRNEGLFVGGSSGANVWGALQAARQL----KKGQKCVVLLPDSSR 317
Query 61 NYTSKFVCDEWMAEKGF 77
NY SKF+ DEWMAE GF
Sbjct 318 NYMSKFISDEWMAEHGF 334
> dre:266987 cbsb, cb442, cbs, wu:fb37g05, wu:fm61c07, wu:fq06c06,
zgc:113704; cystathionine-beta-synthase b (EC:4.2.1.22);
K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=597
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 52/82 (63%), Gaps = 4/82 (4%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K ND ES SR+LIR EG+L GGSSG A++ A+ + + +R ++LPD RN
Sbjct 360 KSNDEESFAMSRMLIREEGLLCGGSSGTAMSAAVNVAKEL----KEGQRCVVILPDSIRN 415
Query 62 YTSKFVCDEWMAEKGFLERSEL 83
Y SKF+ D+WM EKGF+ +L
Sbjct 416 YMSKFLSDKWMFEKGFMSEEDL 437
> hsa:875 CBS, HIP4; cystathionine-beta-synthase (EC:4.2.1.22);
K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=551
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 35/85 (41%), Positives = 55/85 (64%), Gaps = 4/85 (4%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K ND E+ +R+LI EG+L GGS+G+ +A A+KA + + +R ++LPD RN
Sbjct 325 KSNDEEAFTFARMLIAQEGLLCGGSAGSTVAVAVKAAQEL----QEGQRCVVILPDSVRN 380
Query 62 YTSKFVCDEWMAEKGFLERSELSRQ 86
Y +KF+ D WM +KGFL+ +L+ +
Sbjct 381 YMTKFLSDRWMLQKGFLKEEDLTEK 405
> dre:550524 cbsa, MGC174292, zgc:110199; cystathionine-beta-synthase
a; K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=571
Score = 75.5 bits (184), Expect = 3e-14, Method: Composition-based stats.
Identities = 37/82 (45%), Positives = 52/82 (63%), Gaps = 4/82 (4%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K D ES +R+LIR EG+L GGSSG A+A A+K + + +R ++LPD RN
Sbjct 336 KSKDEESFAMARMLIRDEGLLCGGSSGTAMAAAVKFAKEL----KEGQRCVVILPDSIRN 391
Query 62 YTSKFVCDEWMAEKGFLERSEL 83
Y SKF+ D+WM +KGFL ++
Sbjct 392 YMSKFLSDKWMFQKGFLRVEDI 413
> xla:446655 cbs-b, MGC82640, cbs, hip4; cystathionine-beta-synthase
(EC:4.2.1.22); K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=562
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 54/82 (65%), Gaps = 4/82 (4%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K ND ES +R LIR EG+L GGSSG+A++ A+KA + + +R ++LPD RN
Sbjct 318 KSNDEESFFMARALIREEGLLCGGSSGSAMSVAVKAAKDLS----EGQRCVVILPDSVRN 373
Query 62 YTSKFVCDEWMAEKGFLERSEL 83
Y SKF+ D+WM +K FL+ +L
Sbjct 374 YMSKFLSDKWMTQKQFLKVEKL 395
> mmu:12411 Cbs, AI047524, AI303044, HIP4, MGC18856, MGC18895,
MGC37300; cystathionine beta-synthase (EC:4.2.1.22); K01697
cystathionine beta-synthase [EC:4.2.1.22]
Length=561
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 39/83 (46%), Positives = 55/83 (66%), Gaps = 5/83 (6%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K ND +S +R+LI EG+L GGSSG+A+A A+KA + +R ++LPD RN
Sbjct 322 KSNDEDSFAFARMLIAQEGLLCGGSSGSAMAVAVKAAREL----QEGQRCVVILPDSVRN 377
Query 62 YTSKFVCDEWMAEKGFLERSELS 84
Y SKF+ D+WM +KGF+ + ELS
Sbjct 378 YMSKFLSDKWMLQKGFM-KEELS 399
> cel:F54A3.4 hypothetical protein; K01697 cystathionine beta-synthase
[EC:4.2.1.22]
Length=755
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 34/78 (43%), Positives = 53/78 (67%), Gaps = 2/78 (2%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
+K +D ES L +R +IR EG+L GGSSG A+ AL+ ++ +D++ + ++LPDG R
Sbjct 587 LKSDDKESFLMAREIIRTEGILCGGSSGCAVHYALEQCRKLDLPEDAN--VVVLLPDGIR 644
Query 61 NYTSKFVCDEWMAEKGFL 78
NY +KF+ D+WM +GF
Sbjct 645 NYLTKFLDDDWMKARGFF 662
Score = 38.1 bits (87), Expect = 0.006, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 9/77 (11%)
Query 1 VKVNDA-ESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGS 59
V++ DA ES +R LI EG++ G SSGAA+ A+K + + + + +VL DG
Sbjct 229 VEIRDAPESYTFTRHLIETEGIMAGPSSGAAVLEAIKLAKDL----PAGSVVVVVLMDGI 284
Query 60 RNYTSKFVCDEWMAEKG 76
R+Y +WM G
Sbjct 285 RDYLDA----DWMKVNG 297
> cel:ZC373.1 hypothetical protein; K01697 cystathionine beta-synthase
[EC:4.2.1.22]
Length=704
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 35/78 (44%), Positives = 53/78 (67%), Gaps = 2/78 (2%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K +D ES L +R LIR EG+L GGSSG A+ AL+ + + +++ + ++LPDG RN
Sbjct 628 KSHDKESFLMARELIRSEGILCGGSSGCAVHYALEECKSLNLPAEAN--VVVLLPDGIRN 685
Query 62 YTSKFVCDEWMAEKGFLE 79
Y +KF+ D+WM E+ FL+
Sbjct 686 YITKFLDDDWMNERHFLD 703
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 41/71 (57%), Gaps = 4/71 (5%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
+V + E+ +R LI EG++ G SSGAA+ A+K + + + I +VL DG RN
Sbjct 250 EVEEDEAYSFTRHLIGTEGIMAGPSSGAAVLEAIKLAKEL----PAGSTIVVVLMDGIRN 305
Query 62 YTSKFVCDEWM 72
Y F+ D+W+
Sbjct 306 YLRHFLDDDWI 316
> xla:380308 cbs-a, MGC52737, hip4; cystathionine-beta-synthase
(EC:4.2.1.22); K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=565
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 40/82 (48%), Positives = 55/82 (67%), Gaps = 4/82 (4%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
K ND ES +R LIR EG+L GGSSG+A++ A+KA + +G +R ++LPD RN
Sbjct 321 KSNDEESFFMARALIREEGLLCGGSSGSAMSAAMKAAKELG----EGQRCVVILPDSVRN 376
Query 62 YTSKFVCDEWMAEKGFLERSEL 83
Y SKF+ D+WM +K FL+ EL
Sbjct 377 YMSKFLSDKWMTQKQFLKVEEL 398
> sce:YGR155W CYS4, NHS5, STR4, VMA41; Cys4p (EC:4.2.1.22); K01697
cystathionine beta-synthase [EC:4.2.1.22]
Length=507
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 33/86 (38%), Positives = 46/86 (53%), Gaps = 5/86 (5%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMG--WKDDSSKRIAMVLPDGS 59
K +D S +R LI EG+LVGGSSG+A +K E +DD I + PD
Sbjct 265 KTDDKPSFKYARQLISNEGVLVGGSSGSAFTAVVKYCEDHPELTEDDV---IVAIFPDSI 321
Query 60 RNYTSKFVCDEWMAEKGFLERSELSR 85
R+Y +KFV DEW+ + + L+R
Sbjct 322 RSYLTKFVDDEWLKKNNLWDDDVLAR 347
> eco:b2414 cysK, cysZ, ECK2409, JW2407; cysteine synthase A,
O-acetylserine sulfhydrolase A subunit (EC:2.5.1.47); K01738
cysteine synthase A [EC:2.5.1.47]
Length=323
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 41/75 (54%), Gaps = 3/75 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
+ + + E++ +R L+ EG+L G SSGAA+A ALK E + ++K I ++LP
Sbjct 248 IGITNEEAISTARRLMEEEGILAGISSGAAVAAALKLQEDESF---TNKNIVVILPSSGE 304
Query 61 NYTSKFVCDEWMAEK 75
Y S + + EK
Sbjct 305 RYLSTALFADLFTEK 319
> tgo:TGME49_078910 O-acetylserine (thiol) lyase, putative (EC:2.5.1.47);
K01738 cysteine synthase A [EC:2.5.1.47]
Length=499
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 39/76 (51%), Gaps = 4/76 (5%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
+++D ESL + L+ EG+ VG SSG +A A++ +R+G RI +L D
Sbjct 380 EISDVESLTWTHSLLEDEGLAVGMSSGVNVAAAVRLAKRLG----PGHRICTILCDSGTR 435
Query 62 YTSKFVCDEWMAEKGF 77
Y K ++ E+G
Sbjct 436 YAKKQWNISFLREEGL 451
> ath:AT5G28020 CYSD2; CYSD2 (CYSTEINE SYNTHASE D2); catalytic/
cysteine synthase/ pyridoxal phosphate binding (EC:2.5.1.47);
K01738 cysteine synthase A [EC:2.5.1.47]
Length=323
Score = 37.0 bits (84), Expect = 0.017, Method: Composition-based stats.
Identities = 27/69 (39%), Positives = 40/69 (57%), Gaps = 3/69 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
++V E++ ++LL EG+LVG SSGAA A ALK +R +++ K I +V P G
Sbjct 246 IQVAGEEAIETAKLLALKEGLLVGISSGAAAAAALKVAKR---PENAGKLIVVVFPSGGE 302
Query 61 NYTSKFVCD 69
Y S + D
Sbjct 303 RYLSTKLFD 311
> ath:AT3G04940 CYSD1; CYSD1 (CYSTEINE SYNTHASE D1); cysteine
synthase (EC:2.5.1.47); K01738 cysteine synthase A [EC:2.5.1.47]
Length=324
Score = 36.6 bits (83), Expect = 0.020, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 38/64 (59%), Gaps = 3/64 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
++V E++ ++LL EG+LVG SSGAA A A+K +R +++ K I ++ P G
Sbjct 247 IQVTSVEAIETAKLLALKEGLLVGISSGAAAAAAIKVAKR---PENAGKLIVVIFPSGGE 303
Query 61 NYTS 64
Y S
Sbjct 304 RYLS 307
> ath:AT2G43750 OASB; OASB (O-ACETYLSERINE (THIOL) LYASE B); cysteine
synthase; K01738 cysteine synthase A [EC:2.5.1.47]
Length=392
Score = 31.6 bits (70), Expect = 0.69, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 37/64 (57%), Gaps = 3/64 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
+ ++ E++ S+ L EG+LVG SSGAA A A++ +R +++ K IA+V P
Sbjct 314 IAISSEEAIETSKQLALQEGLLVGISSGAAAAAAIQVAKR---PENAGKLIAVVFPSFGE 370
Query 61 NYTS 64
Y S
Sbjct 371 RYLS 374
> cel:C17G1.7 hypothetical protein; K01738 cysteine synthase A
[EC:2.5.1.47]
Length=341
Score = 31.6 bits (70), Expect = 0.70, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPD-GS 59
++++ E+++ ++ L EG+L G SSGA +A AL+ R + + K I LP G
Sbjct 246 IRIHSDEAIVMAQRLSYEEGLLGGISSGANVAAALQLAAR---PEMAGKLIVTCLPSCGE 302
Query 60 RNYTSKFVCD 69
R TS D
Sbjct 303 RYMTSPLYTD 312
> ath:AT4G14880 OASA1; OASA1 (O-ACETYLSERINE (THIOL) LYASE (OAS-TL)
ISOFORM A1); cysteine synthase (EC:2.5.1.47); K01738 cysteine
synthase A [EC:2.5.1.47]
Length=322
Score = 31.2 bits (69), Expect = 0.77, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 37/69 (53%), Gaps = 3/69 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
V+V+ ES+ +R L EG+LVG SSGAA A A+K +R +++ K + P
Sbjct 244 VQVSSDESIDMARQLALKEGLLVGISSGAAAAAAIKLAQR---PENAGKLFVAIFPSFGE 300
Query 61 NYTSKFVCD 69
Y S + D
Sbjct 301 RYLSTVLFD 309
> xla:397727 sox13, Sry, sox12, xSox12, xsox13; SRY (sex determining
region Y)-box 13; K09269 transcription factor SOX5/6/13
(SOX group D)
Length=567
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 32/74 (43%), Gaps = 8/74 (10%)
Query 21 MLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMA------E 74
M + GS G + + A M W D ++I PD + SK + W + +
Sbjct 382 MDIDGSRGNHIKRPMNAF--MVWAKDERRKILQAFPDMHNSSISKILGSRWKSMSNGEKQ 439
Query 75 KGFLERSELSRQLL 88
+ E++ LSRQ L
Sbjct 440 PYYEEQARLSRQHL 453
> ath:AT3G59760 OASC; OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM
C); ATP binding / cysteine synthase (EC:2.5.1.47); K01738
cysteine synthase A [EC:2.5.1.47]
Length=430
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 37/64 (57%), Gaps = 3/64 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
+ ++ E++ ++ L EG++VG SSGAA A A+K +R +++ K IA+V P
Sbjct 352 IAISSEEAIETAKQLALKEGLMVGISSGAAAAAAIKVAKR---PENAGKLIAVVFPSFGE 408
Query 61 NYTS 64
Y S
Sbjct 409 RYLS 412
> mmu:20671 Sox17, Sox; SRY-box containing gene 17; K04495 transcription
factor SOX17 (SOX group F)
Length=419
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 24/84 (28%), Positives = 36/84 (42%), Gaps = 14/84 (16%)
Query 16 IRLEGMLVGGSSG-AALAGALKAIER--------MGWKDDSSKRIAMVLPDGSRNYTSKF 66
++++G +V S A +G KA R M W D KR+A PD SK
Sbjct 42 VKVKGEVVASSGAPAGTSGRAKAESRIRRPMNAFMVWAKDERKRLAQQNPDLHNAELSKM 101
Query 67 VCDEWMA-----EKGFLERSELSR 85
+ W A ++ F+E +E R
Sbjct 102 LGKSWKALTLAEKRPFVEEAERLR 125
> ath:AT5G28030 cysteine synthase, putative / O-acetylserine (thiol)-lyase,
putative / O-acetylserine sulfhydrylase, putative
(EC:2.5.1.47); K01738 cysteine synthase A [EC:2.5.1.47]
Length=323
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 37/64 (57%), Gaps = 3/64 (4%)
Query 1 VKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSR 60
++V E++ ++LL EG+LVG SSGA+ A ALK +R ++ K I ++ P G
Sbjct 246 IQVTGEEAIETTKLLAIKEGLLVGISSGASAAAALKVAKR---PENVGKLIVVIFPSGGE 302
Query 61 NYTS 64
Y S
Sbjct 303 RYLS 306
> ath:AT3G03630 CS26; CS26; cysteine synthase; K01738 cysteine
synthase A [EC:2.5.1.47]
Length=404
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 3/63 (4%)
Query 2 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 61
KV++ E++ +R L EG+LVG SSGAA A+ +R +++ K I ++ P
Sbjct 329 KVSNGEAIEMARRLALEEGLLVGISSGAAAVAAVSLAKR---AENAGKLITVLFPSHGER 385
Query 62 YTS 64
Y +
Sbjct 386 YIT 388
> dre:793481 sterile alpha motif domain containing 9-like
Length=1092
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 25/53 (47%), Gaps = 10/53 (18%)
Query 32 AGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELS 84
AG+LK + W V +G N ++ D WM EKGFL +++S
Sbjct 321 AGSLKLAKSRSW----------VFCNGGINGETQSDDDNWMTEKGFLVENKVS 363
Lambda K H
0.318 0.134 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017039696
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40