bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6482_orf1
Length=127
Score E
Sequences producing significant alignments: (Bits) Value
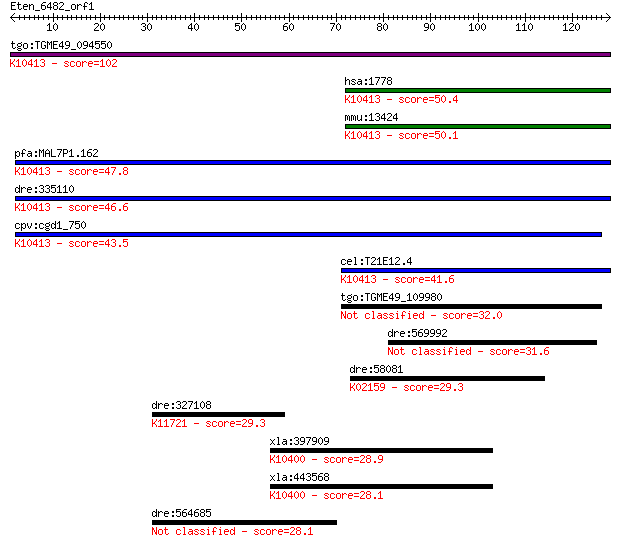
tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein... 102 3e-22
hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNE... 50.4 1e-06
mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,... 50.1 2e-06
pfa:MAL7P1.162 dynein heavy chain, putative; K10413 dynein hea... 47.8 9e-06
dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic 1... 46.6 2e-05
cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain 1,... 43.5 2e-04
cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1); ... 41.6 6e-04
tgo:TGME49_109980 dynein-1-alpha heavy chain, flagellar inner ... 32.0 0.53
dre:569992 dynein, axonemal, heavy chain 5-like 31.6 0.62
dre:58081 baxa, bax, fj16e01, wu:fc50b10, wu:fj16e01; bcl2-ass... 29.3 3.1
dre:327108 brd3b, fa28f05, wu:fa28f05, wu:fc56a11, wu:fj66d12,... 29.3 3.3
xla:397909 kif15-a, XKlp2, kif15, klp2; kinesin family member ... 28.9 4.0
xla:443568 kif15-b, MGC114602, XKlp2, klp2; kinesin family mem... 28.1 6.5
dre:564685 novel protein similar to vertebrate NGFI-A binding ... 28.1 7.1
> tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein
heavy chain 1, cytosolic
Length=4937
Score = 102 bits (254), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 54/131 (41%), Positives = 75/131 (57%), Gaps = 11/131 (8%)
Query 1 QVHPAVEAARQLWMGSLHRIMASVCLLPRFNSRNSARPLKFGGEPTAWDYEDS----EDE 56
Q+ P VEAA Q WMG LH +ASVCL+PR +SR G D+ DS +++
Sbjct 992 QLFPPVEAAHQYWMGELHNTIASVCLVPRLSSR-------IGASAVEDDFADSCSSDDED 1044
Query 57 ALENGDPMKPKSGGTYRAIASRIPPEVIDKAHSAIDRVMGNVEEYVANWMHYQVLWDVDV 116
A E K TYR ++ + P V A+ I++ + V+EYV W+ YQVLWDVDV
Sbjct 1045 AEEKLKKSKLCKNRTYRHLSQLVDPAVFFHAYDVINKHIQRVKEYVQTWLQYQVLWDVDV 1104
Query 117 SEVLGKLGDDI 127
+EV+ ++ DDI
Sbjct 1105 NEVISRVSDDI 1115
> hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNECL,
DYHC, Dnchc1, HL-3, KIAA0325, p22; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4646
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 37/58 (63%), Gaps = 2/58 (3%)
Query 72 YRAIASRIP--PEVIDKAHSAIDRVMGNVEEYVANWMHYQVLWDVDVSEVLGKLGDDI 127
YR +R+P P +++++SA+ ++ VE+YV W+ YQ LWD+ + +LG+D+
Sbjct 1019 YRNALTRMPDGPVALEESYSAVMGIVSEVEQYVKVWLQYQCLWDMQAENIYNRLGEDL 1076
> mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,
Dnchc1, Dnec1, Dnecl, Loa, MAP1C, P22, Swl, mKIAA0325; dynein
cytoplasmic 1 heavy chain 1; K10413 dynein heavy chain
1, cytosolic
Length=4644
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 37/58 (63%), Gaps = 2/58 (3%)
Query 72 YRAIASRIP--PEVIDKAHSAIDRVMGNVEEYVANWMHYQVLWDVDVSEVLGKLGDDI 127
YR +R+P P +++++SA+ ++ VE+YV W+ YQ LWD+ + +LG+D+
Sbjct 1017 YRNALTRMPDGPVALEESYSAVMGIVTEVEQYVKVWLQYQCLWDMQAENIYNRLGEDL 1074
> pfa:MAL7P1.162 dynein heavy chain, putative; K10413 dynein heavy
chain 1, cytosolic
Length=4985
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 30/153 (19%), Positives = 64/153 (41%), Gaps = 29/153 (18%)
Query 2 VHPAVEAARQLWMGSLHRIMASVCLLPRFNS-------RNSARPLKFGGEPTAWDYEDSE 54
+HP++++ RQ+W L + +C + R + N +K G+ T + E +
Sbjct 978 LHPSIDSMRQIWFSKLSDAINIICGITRIKNIYQKKEKTNEKIQIKSKGKNT--NNEKDD 1035
Query 55 DEALENGDPMKPKS--------------------GGTYRAIASRIPPEVIDKAHSAIDRV 94
D D + TY+ I I ++ D A +I+ +
Sbjct 1036 DYIYYTNDSNNKNNIYKNKCDDNNNNIVINDVILDNTYKYIIYFIDKKIYDNAIKSINDM 1095
Query 95 MGNVEEYVANWMHYQVLWDVDVSEVLGKLGDDI 127
+ ++Y + W+ Y+ LW +++ +++ G+DI
Sbjct 1096 VDKAQKYESIWLQYKTLWQIEIGDIISSFGEDI 1128
> dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4643
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/128 (24%), Positives = 55/128 (42%), Gaps = 24/128 (18%)
Query 2 VHPAVEAARQLWMGSLHRIMASVCLLPRFNSRNSARPLKFGGEPTAWDYEDSEDEALENG 61
++P +E R + ++ LPR S+ YE SE+E
Sbjct 970 LNPPIEDCRYRLYQEMFAWKMNILSLPRIQSQRYQ---------VGVHYELSEEEKF--- 1017
Query 62 DPMKPKSGGTYRAIASRIP--PEVIDKAHSAIDRVMGNVEEYVANWMHYQVLWDVDVSEV 119
YR +R+P P +++A++A+ + VE+YV W+ YQ LWD+ +
Sbjct 1018 ----------YRNALTRMPDGPSALEEAYNAVKENVSEVEQYVKVWLQYQCLWDMQPENI 1067
Query 120 LGKLGDDI 127
+L +D+
Sbjct 1068 YNRLEEDL 1075
> cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain
1, cytosolic
Length=5246
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 32/145 (22%), Positives = 65/145 (44%), Gaps = 25/145 (17%)
Query 2 VHPAVEAARQLWMGSLHRIMASVCLLPR-------------FNSRNSAR---PLKFGGEP 45
++P++E + LW+ H+ + + LPR +N + A+ G+
Sbjct 1037 LNPSLEDVKSLWLRRFHQEITKITNLPRVTNISSIPQTSPFYNDISKAKEELATLISGKN 1096
Query 46 TAWD-----YEDSEDEALENGDPMKPKSGGTYRAIASRIPPEVIDKAHSAIDRVMGNVEE 100
D Y +S+D P+ TYR + +P +++ ++ +ID V +++
Sbjct 1097 EQIDENPVNYVNSDDIGSVKHIPLD----HTYRHLFLFLPKSLLEWSYKSIDDVFSSIKV 1152
Query 101 YVANWMHYQVLWDVDVSEVLGKLGD 125
+ W ++ LW V++S V GKL +
Sbjct 1153 SIDYWTRFEALWFVELSSVTGKLNN 1177
> cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1);
K10413 dynein heavy chain 1, cytosolic
Length=4568
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 71 TYRAIASRIP--PEVIDKAHSAIDRVMGNVEEYVANWMHYQVLWDVDVSEVLGKLGDDI 127
TY I + +P ++KA+ ++ +M ++EEY++ W+ YQ LW + ++ LG +
Sbjct 985 TYHNILNVMPEGQACLEKAYDCVNGIMSDLEEYLSEWLSYQSLWVLQAEQLFEMLGTSL 1043
> tgo:TGME49_109980 dynein-1-alpha heavy chain, flagellar inner
arm I1 complex, putative
Length=4629
Score = 32.0 bits (71), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 30/61 (49%), Gaps = 7/61 (11%)
Query 71 TYRAIASRIPPEVIDKA---HSAIDRVMGNVEEYVANWMHYQV---LWDVDVSEVLGKLG 124
T+ + SRIP +ID H AI +V ++ +Y +W Y LWD+ L KL
Sbjct 1088 TFYSEISRIPA-LIDTTVATHQAIQKVFQSISKYARSWKKYDTQWELWDLKRKADLEKLV 1146
Query 125 D 125
D
Sbjct 1147 D 1147
> dre:569992 dynein, axonemal, heavy chain 5-like
Length=4559
Score = 31.6 bits (70), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 27/47 (57%), Gaps = 3/47 (6%)
Query 81 PEVIDKAHS---AIDRVMGNVEEYVANWMHYQVLWDVDVSEVLGKLG 124
P++ ++A + I R++ + Y+ +WM Y+ LW +D + V+ K
Sbjct 1055 PQINEQATAVSQTIQRLLNGISVYLKSWMSYRSLWKLDKAIVMEKFA 1101
> dre:58081 baxa, bax, fj16e01, wu:fc50b10, wu:fj16e01; bcl2-associated
X protein, a; K02159 apoptosis regulator BAX, membrane
protein (BCL2-associated)
Length=192
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 4/41 (9%)
Query 73 RAIASRIPPEVIDKAHSAIDRVMGNVEEYVANWMHYQVLWD 113
+AI++R+P D + I M ++E+V NW+ Q WD
Sbjct 125 KAISTRVP----DIIRTIISWTMSYIQEHVINWIREQGGWD 161
> dre:327108 brd3b, fa28f05, wu:fa28f05, wu:fc56a11, wu:fj66d12,
zgc:77289; bromodomain containing 3b; K11721 bromodomain-containing
protein 3
Length=664
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 31 NSRNSARPLKFGGEPTAWDYEDSEDEAL 58
NS ++ R K GG+P A +YE E+E+L
Sbjct 523 NSTSTTRQAKKGGKPGAANYESDEEESL 550
> xla:397909 kif15-a, XKlp2, kif15, klp2; kinesin family member
15; K10400 kinesin family member 15
Length=1388
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query 56 EALENGDPMKPKSGGTYRAIASR-IPPEVIDKAHSAIDRVMGNVEEYV 102
+A+ NG PKSG I + IPPE+ ++A+ AI + V+E V
Sbjct 685 DAMPNGLMDTPKSGDVMDDIINEPIPPEMSEQAYEAIAEELRIVQEQV 732
> xla:443568 kif15-b, MGC114602, XKlp2, klp2; kinesin family member
15; K10400 kinesin family member 15
Length=1387
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query 56 EALENGDPMKPKSGGTYRAIASR-IPPEVIDKAHSAIDRVMGNVEEYV 102
+ + NG PKSG I + IPPE+ ++A+ AI + V+E V
Sbjct 683 DGISNGLTDTPKSGDVMDDIINEPIPPEMSEQAYEAIAEELRIVQEQV 730
> dre:564685 novel protein similar to vertebrate NGFI-A binding
protein 1 (EGR1 binding protein 1) (NAB1)
Length=500
Score = 28.1 bits (61), Expect = 7.1, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 19/43 (44%), Gaps = 4/43 (9%)
Query 31 NSRNSARPLKFGGEPTAWDYEDSEDE----ALENGDPMKPKSG 69
NS + PL+ G EP W +E E E G P P+ G
Sbjct 140 NSSATVSPLQSGSEPRFWANHGTESEHSLSPAELGSPSSPRDG 182
Lambda K H
0.317 0.134 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2054672932
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40