bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6470_orf2
Length=70
Score E
Sequences producing significant alignments: (Bits) Value
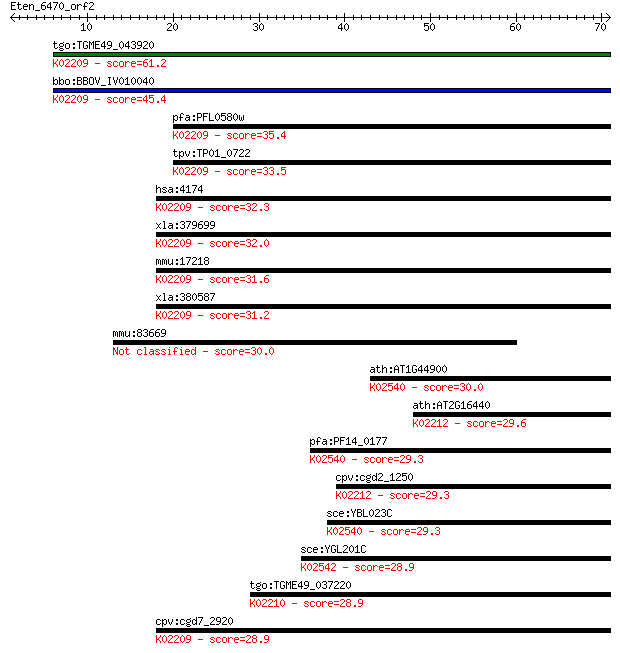
tgo:TGME49_043920 DNA replication licensing factor, putative ;... 61.2 7e-10
bbo:BBOV_IV010040 23.m06024; DNA replication licensing factor ... 45.4 4e-05
pfa:PFL0580w DNA replication licensing factor MCM5, putative; ... 35.4 0.049
tpv:TP01_0722 DNA replication licensing factor MCM5; K02209 mi... 33.5 0.16
hsa:4174 MCM5, CDC46, MGC5315, P1-CDC46; minichromosome mainte... 32.3 0.34
xla:379699 mcm5-b, MGC68977, cdc46, xmcm5; minichromosome main... 32.0 0.51
mmu:17218 Mcm5, AA617332, AI324988, AL033333, Cdc46, Mcmd5, P1... 31.6 0.60
xla:380587 mcm5-a, MGC53425, cdc46, mcm5, xmcm5; minichromosom... 31.2 0.78
mmu:83669 Wdr6, mWDR6; WD repeat domain 6 30.0
ath:AT1G44900 ATP binding / DNA binding / DNA-dependent ATPase... 30.0 1.8
ath:AT2G16440 DNA replication licensing factor, putative; K022... 29.6 2.2
pfa:PF14_0177 DNA replication licensing factor MCM2; K02540 mi... 29.3 2.8
cpv:cgd2_1250 DNA replication licensing factor MCM4 like AAA+ ... 29.3 3.4
sce:YBL023C MCM2; Mcm2p; K02540 minichromosome maintenance pro... 29.3 3.5
sce:YGL201C MCM6; Mcm6p; K02542 minichromosome maintenance pro... 28.9 4.1
tgo:TGME49_037220 DNA replication licensing factor, putative (... 28.9 4.3
cpv:cgd7_2920 DNA replication licensing factor MCM5 like AAA+ ... 28.9 4.6
> tgo:TGME49_043920 DNA replication licensing factor, putative
; K02209 minichromosome maintenance protein 5 (cell division
control protein 46)
Length=794
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 30/68 (44%), Positives = 46/68 (67%), Gaps = 5/68 (7%)
Query 6 NVAAERYDQSVPQQAYIHVLGIKKINFSPLLST---LTSLEQRSLFQRLSQEPNIIDKIA 62
+ ++ DQ P +Y+HVLG++K++F L T T EQR +F +L+Q NI+DKIA
Sbjct 345 HTGSKETDQ--PHSSYLHVLGLQKVSFMSLARTRLSFTQTEQRDVFYKLAQSQNILDKIA 402
Query 63 RSIAPALF 70
+S+APAL+
Sbjct 403 KSLAPALY 410
> bbo:BBOV_IV010040 23.m06024; DNA replication licensing factor
MCM5; K02209 minichromosome maintenance protein 5 (cell division
control protein 46)
Length=777
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 0/65 (0%)
Query 6 NVAAERYDQSVPQQAYIHVLGIKKINFSPLLSTLTSLEQRSLFQRLSQEPNIIDKIARSI 65
NV A D + +Y+HVLGI+K+ + LE+ + L+ +P+I DKI RSI
Sbjct 331 NVNARGNDSTAIGSSYLHVLGIEKLTQGKGEAISFDLEETNDLVLLATQPDIHDKIFRSI 390
Query 66 APALF 70
APA++
Sbjct 391 APAIY 395
> pfa:PFL0580w DNA replication licensing factor MCM5, putative;
K02209 minichromosome maintenance protein 5 (cell division
control protein 46)
Length=758
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 20 AYIHVLGIKKINFSPLLSTLTSLEQRSLFQRLSQEPNIIDKIARSIAPALF 70
+Y+HVLG +K + +E+R+ L+ E NI +KI +SIAP L+
Sbjct 328 SYLHVLGFQKYDDMSGNDLNFDVEERNELTLLAAEHNIHEKIFKSIAPELY 378
> tpv:TP01_0722 DNA replication licensing factor MCM5; K02209
minichromosome maintenance protein 5 (cell division control
protein 46)
Length=767
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 32/51 (62%), Gaps = 2/51 (3%)
Query 20 AYIHVLGIKKINFSPLLSTLTSLEQRSLFQRLSQEPNIIDKIARSIAPALF 70
+Y+HVLGI+K+N + +++ + L+ +P+I KI SIAP+++
Sbjct 337 SYLHVLGIQKLNVNETYE--FDIDEGNDLLLLASQPDIHTKIFNSIAPSIY 385
> hsa:4174 MCM5, CDC46, MGC5315, P1-CDC46; minichromosome maintenance
complex component 5; K02209 minichromosome maintenance
protein 5 (cell division control protein 46)
Length=734
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 32/55 (58%), Gaps = 2/55 (3%)
Query 18 QQAYIHVLGIK--KINFSPLLSTLTSLEQRSLFQRLSQEPNIIDKIARSIAPALF 70
+ +YI VLGI+ + S ++ F+RL+ PN+ + I++SIAP++F
Sbjct 290 RSSYIRVLGIQVDTDGSGRSFAGAVSPQEEEEFRRLAALPNVYEVISKSIAPSIF 344
> xla:379699 mcm5-b, MGC68977, cdc46, xmcm5; minichromosome maintenance
complex component 5 (EC:3.6.4.12); K02209 minichromosome
maintenance protein 5 (cell division control protein
46)
Length=735
Score = 32.0 bits (71), Expect = 0.51, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 35/56 (62%), Gaps = 4/56 (7%)
Query 18 QQAYIHVLGIK---KINFSPLLSTLTSLEQRSLFQRLSQEPNIIDKIARSIAPALF 70
+ +YI V+GI+ + T+T E+ F+RL+ +P+I + +A+SIAP+++
Sbjct 291 RSSYIRVVGIQVDTEGTGRSAAGTITPQEEEE-FRRLAVKPDIYETVAKSIAPSIY 345
> mmu:17218 Mcm5, AA617332, AI324988, AL033333, Cdc46, Mcmd5,
P1-CDC46, mCD46, mCDC46; minichromosome maintenance deficient
5, cell division cycle 46 (S. cerevisiae) (EC:3.6.4.12); K02209
minichromosome maintenance protein 5 (cell division control
protein 46)
Length=734
Score = 31.6 bits (70), Expect = 0.60, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 33/55 (60%), Gaps = 2/55 (3%)
Query 18 QQAYIHVLGIKKINFSPLLSTLTSL--EQRSLFQRLSQEPNIIDKIARSIAPALF 70
+ +YI VLGI+ S S+ ++ F+RL+ PNI + I++SI+P++F
Sbjct 290 RSSYIRVLGIQVDTDGSGRSFAGSVSPQEEEEFRRLAALPNIYELISKSISPSIF 344
> xla:380587 mcm5-a, MGC53425, cdc46, mcm5, xmcm5; minichromosome
maintenance complex component 5 (EC:3.6.4.12); K02209 minichromosome
maintenance protein 5 (cell division control protein
46)
Length=735
Score = 31.2 bits (69), Expect = 0.78, Method: Composition-based stats.
Identities = 16/55 (29%), Positives = 34/55 (61%), Gaps = 2/55 (3%)
Query 18 QQAYIHVLGIKKINFSPLLSTLTSL--EQRSLFQRLSQEPNIIDKIARSIAPALF 70
+ +YI V+GI+ S ++ ++ F+RL+ +P+I + +A+SIAP+++
Sbjct 291 RSSYIRVVGIQVDTEGTGRSAAGAITPQEEEEFRRLAAKPDIYETVAKSIAPSIY 345
> mmu:83669 Wdr6, mWDR6; WD repeat domain 6
Length=1125
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 26/48 (54%), Gaps = 3/48 (6%)
Query 13 DQSVPQQAYIHVLGIKKINFSPLLSTLTSLEQRSLFQRLSQ-EPNIID 59
+ SVP HV G+K + SP L S++QR F RL Q EP ++
Sbjct 1041 EYSVPCAHAAHVTGVKIL--SPKLMVSASIDQRLTFWRLGQGEPTFMN 1086
> ath:AT1G44900 ATP binding / DNA binding / DNA-dependent ATPase;
K02540 minichromosome maintenance protein 2
Length=936
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 10/28 (35%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 43 EQRSLFQRLSQEPNIIDKIARSIAPALF 70
E ++ + LS++P I+++I +SIAP+++
Sbjct 485 EDKTQIEELSKDPRIVERIIKSIAPSIY 512
> ath:AT2G16440 DNA replication licensing factor, putative; K02212
minichromosome maintenance protein 4 (cell division control
protein 54)
Length=847
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 10/23 (43%), Positives = 20/23 (86%), Gaps = 0/23 (0%)
Query 48 FQRLSQEPNIIDKIARSIAPALF 70
FQ LS++P+I ++++RS+AP ++
Sbjct 426 FQELSKQPDIYERLSRSLAPNIW 448
> pfa:PF14_0177 DNA replication licensing factor MCM2; K02540
minichromosome maintenance protein 2
Length=971
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 25/35 (71%), Gaps = 1/35 (2%)
Query 36 LSTLTSLEQRSLFQRLSQEPNIIDKIARSIAPALF 70
LS LT + + + +LS++PNI ++I SIAPA++
Sbjct 466 LSELTEDDIKDIL-KLSKDPNIRERIITSIAPAIW 499
> cpv:cgd2_1250 DNA replication licensing factor MCM4 like AAA+
ATpase ; K02212 minichromosome maintenance protein 4 (cell
division control protein 54)
Length=896
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 27/40 (67%), Gaps = 8/40 (20%)
Query 39 LTSLEQRSLFQR--------LSQEPNIIDKIARSIAPALF 70
+ S+E+ +LF + +S++P + DK++RSIAP+++
Sbjct 424 INSVEKNNLFTKEMIEQFHAMSKDPMLYDKLSRSIAPSIW 463
> sce:YBL023C MCM2; Mcm2p; K02540 minichromosome maintenance protein
2
Length=868
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 24/33 (72%), Gaps = 1/33 (3%)
Query 38 TLTSLEQRSLFQRLSQEPNIIDKIARSIAPALF 70
+ T E+R F+++S++ IIDKI S+AP+++
Sbjct 475 SWTEEEERE-FRKISRDRGIIDKIISSMAPSIY 506
> sce:YGL201C MCM6; Mcm6p; K02542 minichromosome maintenance protein
6
Length=1017
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 25/36 (69%), Gaps = 1/36 (2%)
Query 35 LLSTLTSLEQRSLFQRLSQEPNIIDKIARSIAPALF 70
L++L+S E L + + ++ +I DK+ RSIAPA+F
Sbjct 504 FLNSLSSDEINEL-KEMVKDEHIYDKLVRSIAPAVF 538
> tgo:TGME49_037220 DNA replication licensing factor, putative
(EC:6.6.1.1); K02210 minichromosome maintenance protein 7 (cell
division control protein 47)
Length=865
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 6/48 (12%)
Query 29 KINFSPLLSTLTSLEQRSLFQ------RLSQEPNIIDKIARSIAPALF 70
+++F L T L++R Q +L Q P + D +ARSIAP ++
Sbjct 385 EVSFIQLKKQQTHLDRRESLQIAEKVAKLRQTPGLYDLLARSIAPGVY 432
> cpv:cgd7_2920 DNA replication licensing factor MCM5 like AAA+
ATpase ; K02209 minichromosome maintenance protein 5 (cell
division control protein 46)
Length=791
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 14/65 (21%)
Query 18 QQAYIHVLGIKKINFSPLLS---TLTSLEQRSL---------FQRLSQEPNIIDKIARSI 65
+ +Y++V+G+ +N+ S T T ++ S+ F+R+S PNI + I SI
Sbjct 338 RTSYLYVIGV--MNYGSSWSNKNTNTLIKNSSISNQYNEIEEFRRISSLPNIHELIVNSI 395
Query 66 APALF 70
APA++
Sbjct 396 APAIY 400
Lambda K H
0.317 0.131 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2024947620
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40