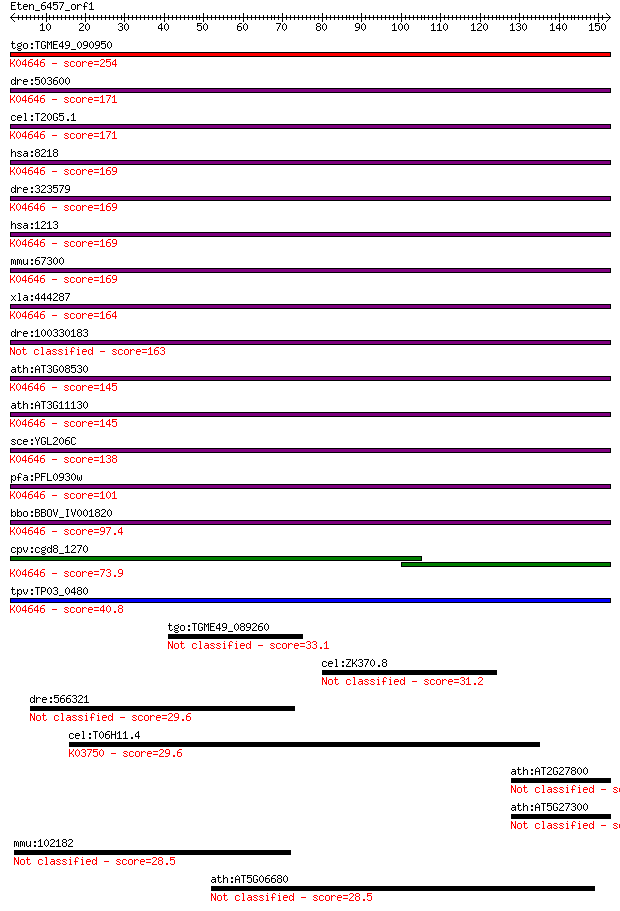
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6457_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_090950 clathrin heavy chain, putative ; K04646 clat... 254 1e-67
dre:503600 cltcb, im:7145213, zgc:113234; clathrin, heavy poly... 171 8e-43
cel:T20G5.1 chc-1; Clathrin Heavy Chain family member (chc-1);... 171 1e-42
hsa:8218 CLTCL1, CHC22, CLH22, CLTCL, CLTD, FLJ36032; clathrin... 169 2e-42
dre:323579 cltca, cb1033, cltc, wu:fc03e11, wu:fc49a11, wu:fd0... 169 3e-42
hsa:1213 CLTC, CHC, CHC17, CLH-17, CLTCL2, Hc, KIAA0034; clath... 169 3e-42
mmu:67300 Cltc, 3110065L21Rik, CHC, MGC92975, R74732; clathrin... 169 3e-42
xla:444287 cltc, MGC80936; clathrin, heavy chain (Hc); K04646 ... 164 1e-40
dre:100330183 cltc; clathrin, heavy chain (Hc) 163 2e-40
ath:AT3G08530 clathrin heavy chain, putative; K04646 clathrin ... 145 3e-35
ath:AT3G11130 clathrin heavy chain, putative; K04646 clathrin ... 145 4e-35
sce:YGL206C CHC1, SWA5; Chc1p; K04646 clathrin heavy chain 138 8e-33
pfa:PFL0930w clathrin heavy chain, putative; K04646 clathrin h... 101 1e-21
bbo:BBOV_IV001820 21.m02773; clathrin heavy chain; K04646 clat... 97.4 1e-20
cpv:cgd8_1270 clathrin heavy chain ; K04646 clathrin heavy chain 73.9 2e-13
tpv:TP03_0480 clathrin heavy chain; K04646 clathrin heavy chain 40.8 0.002
tgo:TGME49_089260 hypothetical protein 33.1 0.35
cel:ZK370.8 hypothetical protein 31.2 1.5
dre:566321 importin-13-like 29.6 4.0
cel:T06H11.4 moc-1; MOlybdenum Cofactor biosynthesis family me... 29.6 4.2
ath:AT2G27800 hypothetical protein 28.9 7.0
ath:AT5G27300 hypothetical protein 28.5 8.4
mmu:102182 Prmt10, AI931714; protein arginine methyltransferas... 28.5 8.7
ath:AT5G06680 SPC98; SPC98 (SPINDLE POLE BODY COMPONENT 98); t... 28.5 8.9
> tgo:TGME49_090950 clathrin heavy chain, putative ; K04646 clathrin
heavy chain
Length=1731
Score = 254 bits (648), Expect = 1e-67, Method: Compositional matrix adjust.
Identities = 119/152 (78%), Positives = 134/152 (88%), Gaps = 0/152 (0%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSS 60
CE GL QRALE YT++AD+KRV+LQ+GGKISQE+ QFFG + PD +EILTDMLRSSS
Sbjct 647 CEKVGLSQRALELYTDIADIKRVMLQSGGKISQEFTQQFFGNLPPDASLEILTDMLRSSS 706
Query 61 QNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAAA 120
QNLQAVV VAIKFH Q+G +L+ MFE+FSSYEG+FYFLGSILAFS+DPEVHFKYIEAAA
Sbjct 707 QNLQAVVAVAIKFHGQIGTTKLVEMFEKFSSYEGVFYFLGSILAFSSDPEVHFKYIEAAA 766
Query 121 KLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
KLNHTQEVERVCRE K YEPQRVKEFLKQ +L
Sbjct 767 KLNHTQEVERVCRESKCYEPQRVKEFLKQVKL 798
> dre:503600 cltcb, im:7145213, zgc:113234; clathrin, heavy polypeptide
b (Hc); K04646 clathrin heavy chain
Length=1677
Score = 171 bits (433), Expect = 8e-43, Method: Compositional matrix adjust.
Identities = 84/153 (54%), Positives = 110/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE+YT+L D+KR ++ T ++ EW+ FFG +S + +E L ML ++
Sbjct 617 CEKAGLLQRALEHYTDLYDIKRAVVHTH-LLNPEWLVNFFGSLSVEDSMECLRAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+HEQL + L +FE F S+EG+FYFLGSI+ FS DPEVHFKYI+AA
Sbjct 676 RQNLQICVQVASKYHEQLSTQSLTELFESFKSFEGLFYFLGSIVNFSQDPEVHFKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y+P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERICRESNCYDPERVKNFLKEAKL 768
> cel:T20G5.1 chc-1; Clathrin Heavy Chain family member (chc-1);
K04646 clathrin heavy chain
Length=1681
Score = 171 bits (432), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 83/153 (54%), Positives = 111/153 (72%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE++T+L D+KR ++ T + +W+ +FG +S + VE L ML +
Sbjct 619 CEKAGLLQRALEHFTDLYDIKRTVVHTH-LLKPDWLVGYFGSLSVEDSVECLKAMLTQNI 677
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ VV +A K+HEQLG ++LI MFE SYEG+FYFLGSI+ FS DPEVHFKYI+AA
Sbjct 678 RQNLQVVVQIASKYHEQLGADKLIEMFENHKSYEGLFYFLGSIVNFSQDPEVHFKYIQAA 737
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
+ +EVER+CRE + Y+ +RVK FLK+A+L
Sbjct 738 TRTGQIKEVERICRESQCYDAERVKNFLKEAKL 770
> hsa:8218 CLTCL1, CHC22, CLH22, CLTCL, CLTD, FLJ36032; clathrin,
heavy chain-like 1; K04646 clathrin heavy chain
Length=1583
Score = 169 bits (429), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 83/153 (54%), Positives = 110/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL Q+ALE+YT+L D+KR ++ T ++ EW+ FFG +S + VE L ML ++
Sbjct 617 CEKAGLLQQALEHYTDLYDIKRAVVHTH-LLNPEWLVNFFGSLSVEDSVECLHAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+HEQLG + L+ +FE F SY+G+FYFLGSI+ FS DP+VH KYI+AA
Sbjct 676 RQNLQLCVQVASKYHEQLGTQALVELFESFKSYKGLFYFLGSIVNFSQDPDVHLKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERICRESSCYNPERVKNFLKEAKL 768
> dre:323579 cltca, cb1033, cltc, wu:fc03e11, wu:fc49a11, wu:fd07f02;
clathrin, heavy polypeptide a (Hc); K04646 clathrin
heavy chain
Length=1680
Score = 169 bits (429), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 83/153 (54%), Positives = 109/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE+YT+L D+KR ++ T ++ EW+ +FG +S + +E L ML ++
Sbjct 617 CEKAGLLQRALEHYTDLYDIKRAVVHTH-LLNPEWLVNYFGSLSVEDSLECLRAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+HEQL + L +FE F S+EG+FYFLGSI+ FS DPEVHFKYI+AA
Sbjct 676 RQNLQICVQVASKYHEQLSTQSLTELFESFKSFEGLFYFLGSIVNFSQDPEVHFKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERICRESNCYNPERVKNFLKEAKL 768
> hsa:1213 CLTC, CHC, CHC17, CLH-17, CLTCL2, Hc, KIAA0034; clathrin,
heavy chain (Hc); K04646 clathrin heavy chain
Length=1675
Score = 169 bits (429), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 82/153 (53%), Positives = 111/153 (72%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE++T+L D+KR ++ T ++ EW+ +FG +S + +E L ML ++
Sbjct 617 CEKAGLLQRALEHFTDLYDIKRAVVHTH-LLNPEWLVNYFGSLSVEDSLECLRAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+HEQL + LI +FE F S+EG+FYFLGSI+ FS DP+VHFKYI+AA
Sbjct 676 RQNLQICVQVASKYHEQLSTQSLIELFESFKSFEGLFYFLGSIVNFSQDPDVHFKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y+P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERICRESNCYDPERVKNFLKEAKL 768
> mmu:67300 Cltc, 3110065L21Rik, CHC, MGC92975, R74732; clathrin,
heavy polypeptide (Hc); K04646 clathrin heavy chain
Length=1675
Score = 169 bits (429), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 82/153 (53%), Positives = 111/153 (72%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE++T+L D+KR ++ T ++ EW+ +FG +S + +E L ML ++
Sbjct 617 CEKAGLLQRALEHFTDLYDIKRAVVHTH-LLNPEWLVNYFGSLSVEDSLECLRAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+HEQL + LI +FE F S+EG+FYFLGSI+ FS DP+VHFKYI+AA
Sbjct 676 RQNLQICVQVASKYHEQLSTQSLIELFESFKSFEGLFYFLGSIVNFSQDPDVHFKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y+P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERICRESNCYDPERVKNFLKEAKL 768
> xla:444287 cltc, MGC80936; clathrin, heavy chain (Hc); K04646
clathrin heavy chain
Length=1675
Score = 164 bits (414), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 81/153 (52%), Positives = 109/153 (71%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE++T+L D+KR ++ T ++ EW+ +FG +S + +E L ML ++
Sbjct 617 CEKAGLLQRALEHFTDLYDIKRAVVHTH-LLNPEWLVNYFGSLSVEDSLECLRAMLSANI 675
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V VA K+HEQL LI +FE F S+EG+FYFLGSI+ FS DP+VHFKYI+AA
Sbjct 676 RQNLQICVQVASKYHEQLSTLSLIELFESFKSFEGLFYFLGSIVNFSQDPDVHFKYIQAA 735
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+ RE Y+P+RVK FLK+A+L
Sbjct 736 CKTGQIKEVERISRESNCYDPERVKNFLKEAKL 768
> dre:100330183 cltc; clathrin, heavy chain (Hc)
Length=951
Score = 163 bits (413), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 79/153 (51%), Positives = 107/153 (69%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSS- 59
CE AGL QRALE+Y++ D+KR ++ T ++ EW+ FFG +S ++ L ML ++
Sbjct 123 CEKAGLLQRALEHYSDPYDIKRAVVHTH-LLNPEWLVNFFGSLSVGDSIDCLKAMLSANL 181
Query 60 SQNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
QNLQ V +A K+HEQLG + L+ +FE F SYEG+FYFLGSI+ FS DP+VHFKYI+AA
Sbjct 182 RQNLQLSVQIATKYHEQLGTQTLVELFESFKSYEGLFYFLGSIVNFSQDPDVHFKYIQAA 241
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
K +EVER+CRE Y + VK FLK+A+L
Sbjct 242 CKTGQIKEVERICRESSCYNAEHVKNFLKEAKL 274
> ath:AT3G08530 clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1703
Score = 145 bits (367), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 71/153 (46%), Positives = 108/153 (70%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSS 60
CE AGLY ++L++Y+EL D+KRV++ T I + + +FFG +S + +E + D+L +
Sbjct 631 CEKAGLYIQSLKHYSELPDIKRVIVNTHA-IEPQALVEFFGTLSSEWAMECMKDLLLVNL 689
Query 61 Q-NLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
+ NLQ +V ++ EQLG++ I +FE+F SYEG+++FLGS L+ S DPE+HFKYIEAA
Sbjct 690 RGNLQIIVQACKEYCEQLGVDACIKLFEQFKSYEGLYFFLGSYLSMSEDPEIHFKYIEAA 749
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
AK +EVERV RE +Y+ ++ K FL +A+L
Sbjct 750 AKTGQIKEVERVTRESNFYDAEKTKNFLMEAKL 782
> ath:AT3G11130 clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1705
Score = 145 bits (367), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 71/153 (46%), Positives = 108/153 (70%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSS 60
CE AGLY ++L++Y+EL D+KRV++ T I + + +FFG +S + +E + D+L +
Sbjct 631 CEKAGLYIQSLKHYSELPDIKRVIVNTHA-IEPQALVEFFGTLSSEWAMECMKDLLLVNL 689
Query 61 Q-NLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
+ NLQ +V ++ EQLG++ I +FE+F SYEG+++FLGS L+ S DPE+HFKYIEAA
Sbjct 690 RGNLQIIVQACKEYCEQLGVDACIKLFEQFKSYEGLYFFLGSYLSMSEDPEIHFKYIEAA 749
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
AK +EVERV RE +Y+ ++ K FL +A+L
Sbjct 750 AKTGQIKEVERVTRESNFYDAEKTKNFLMEAKL 782
> sce:YGL206C CHC1, SWA5; Chc1p; K04646 clathrin heavy chain
Length=1653
Score = 138 bits (347), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 63/153 (41%), Positives = 104/153 (67%), Gaps = 2/153 (1%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSS 60
E AGLYQRALENYT++ D+KR ++ T + +W+ +FG+++ + + L ++ ++
Sbjct 623 SEKAGLYQRALENYTDIKDIKRCVVHTNA-LPIDWLVGYFGKLNVEQSLACLKALMDNNI 681
Query 61 Q-NLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIEAA 119
Q N+Q VV VA KF + +G LI +FE +++ EG++Y+L S++ + D +V +KYIEAA
Sbjct 682 QANIQTVVQVATKFSDLIGPSTLIKLFEDYNATEGLYYYLASLVNLTEDKDVVYKYIEAA 741
Query 120 AKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
AK+ +E+ER+ ++ Y+P+RVK FLK A L
Sbjct 742 AKMKQYREIERIVKDNNVYDPERVKNFLKDANL 774
> pfa:PFL0930w clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1997
Score = 101 bits (251), Expect = 1e-21, Method: Composition-based stats.
Identities = 68/239 (28%), Positives = 110/239 (46%), Gaps = 87/239 (36%)
Query 1 CEAAGLYQRALENYTELADVKRVL-----LQTGG-------------------------- 29
CE GLYQRALENYT + D+KRV+ Q GG
Sbjct 735 CEEKGLYQRALENYTNINDIKRVITKSTCFQKGGNNNNTTTTTTTTSNMGDGHFDMNNVS 794
Query 30 --KISQEWMAQFFGRMSPDICVEILTDMLRSSSQNLQAVVGVAIKFHEQLGMERLIAMFE 87
KIS EW+ +F +S +C E+L D ++ S N++ V+ + +++++++G++++I FE
Sbjct 795 KGKISIEWIKNYFSTLSDSVCEELLFDFMKGSKINMEVVISICVQYYDKIGIKKIINKFE 854
Query 88 RFSSYEGIFYFLGSILA--------------------------------------FSTDP 109
+YEGIFYF+ +IL + P
Sbjct 855 ENKNYEGIFYFVSNILNDLPNLIVKGNNNDNLSNNSLSTYNNMNDEDASILLTSDICSTP 914
Query 110 E------------VHF---KYIEAAAKLNHTQEVERVCRE-GKYYEPQRVKEFLKQAQL 152
+ VH+ KYIEA K+N+ QE++R+C++ Y P+++K FLK +L
Sbjct 915 QYSYETANLKLEDVHYIMFKYIEACVKINNIQELDRICKDKNAKYNPEQIKNFLKDCKL 973
> bbo:BBOV_IV001820 21.m02773; clathrin heavy chain; K04646 clathrin
heavy chain
Length=1676
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 47/161 (29%), Positives = 91/161 (56%), Gaps = 9/161 (5%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSS 60
CE AG++ A+++Y DVKR++++ GG +++ + + MSP+ +E+L +ML S+
Sbjct 649 CEDAGMFDMAIQHYNSFFDVKRLIIKAGGNMNRAVLEKSMKNMSPENALEVLREMLDSAE 708
Query 61 QNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFS---------TDPEV 111
+ VV A+ H +G +++ +FER +S + +F FL ++ S + +
Sbjct 709 ISNDHVVSCALTMHNHIGTMQVVQLFERSASSDVLFSFLRALPVISQGSAAESTEQNATI 768
Query 112 HFKYIEAAAKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
+ +I+ N +++ER+C+E Y+ RVK+ LKQ+ L
Sbjct 769 VYTFIKCCIDRNEMEDLERICKESNVYDGVRVKDLLKQSAL 809
> cpv:cgd8_1270 clathrin heavy chain ; K04646 clathrin heavy chain
Length=2007
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 43/148 (29%), Positives = 73/148 (49%), Gaps = 44/148 (29%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSP----DICVEIL---- 52
CE AGLY+RALEN++++ D++R+L G ++ +W+A + ++SP D E+L
Sbjct 691 CEKAGLYERALENFSDMRDIRRILGVACGSLNTDWLANYLSKLSPRTRFDCLKELLLVCK 750
Query 53 --------------------------TDMLRSSSQN----------LQAVVGVAIKFHEQ 76
++ +SS N LQ+V+ V IK +
Sbjct 751 NSSGAGGLVSMGGGLASNGALPGNGQAGLIGNSSSNSIALSNNNTVLQSVIQVCIKNVDN 810
Query 77 LGMERLIAMFERFSSYEGIFYFLGSILA 104
+G+E +I +FE+ +EGI+Y +GS L
Sbjct 811 IGIENIITLFEQQGIWEGIYYVVGSNLT 838
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 3/54 (5%)
Query 100 GSILAFSTDPEVHFKYIEAAAKLNHTQEVERVCRE-GKYYEPQRVKEFLKQAQL 152
G +FST + FKYIEA+ L QE ER+CR+ + YEP++V E+ K ++
Sbjct 894 GGTGSFST--FIVFKYIEASVHLGQIQEAERICRDFPQSYEPEQVIEYFKSIKM 945
> tpv:TP03_0480 clathrin heavy chain; K04646 clathrin heavy chain
Length=1696
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 36/158 (22%), Positives = 73/158 (46%), Gaps = 11/158 (6%)
Query 1 CEAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSS 60
CE LY+ +L+ YT+L D+KRV+L+ +S+ + + M + +E+L ML +
Sbjct 669 CEQFELYEYSLKFYTKLQDIKRVVLKGINVLSRTTLNKTLLAMGEEDVLELLRAMLENVQ 728
Query 61 QNLQAVVGVAIKFHEQLGMERLIAMFERFSSYEGIFYFLGSILAFSTDP------EVHFK 114
L VV + + ++ +L ++ + FL S+ ++ E+ +
Sbjct 729 SEL--VVSSCLALYGKIDAMKL---YQLLQDNPIAYQFLKSLPIIHSNTVDKDSNELVLR 783
Query 115 YIEAAAKLNHTQEVERVCREGKYYEPQRVKEFLKQAQL 152
YI + E+ER+ + + + KE +K ++L
Sbjct 784 YISLCISKDEITELERLLLQNNNFNLSQAKELIKSSRL 821
> tgo:TGME49_089260 hypothetical protein
Length=158
Score = 33.1 bits (74), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 41 GRMSPDICVEILTDMLRSSSQNLQAVVGVAIKFH 74
GR++P +C+EI+ D++ SS + A + V K H
Sbjct 114 GRVAPGLCLEIIADLVASSPGPISAAIIVTHKAH 147
> cel:ZK370.8 hypothetical protein
Length=569
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 80 ERLIAMFERFSSYEGIFYFLGSILAFSTDPEVHFKYIE-AAAKLN 123
E LI + +SY G +L++STDP +YI+ AK+N
Sbjct 238 EELIDLLTEENSYPPAMILRGKMLSYSTDPNEATRYIDKVGAKIN 282
> dre:566321 importin-13-like
Length=945
Score = 29.6 bits (65), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 37/67 (55%), Gaps = 1/67 (1%)
Query 6 LYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSSQNLQA 65
L+ + +Y +L V+ VL Q GK+ + + + G SP E+L ++L S S+N +
Sbjct 822 LFTELISHYEDLPTVRDVL-QEDGKLLLQTLLEAIGGQSPRSLAELLAEVLFSVSRNCPS 880
Query 66 VVGVAIK 72
++ + ++
Sbjct 881 LLTLWLR 887
> cel:T06H11.4 moc-1; MOlybdenum Cofactor biosynthesis family
member (moc-1); K03750 molybdopterin biosynthesis protein MoeA
Length=435
Score = 29.6 bits (65), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 27/138 (19%), Positives = 62/138 (44%), Gaps = 30/138 (21%)
Query 16 ELADVKRVLLQTGGKISQ-------EWMAQFFGRMSPDICVEILTDMLRSSSQNLQAVV- 67
EL + ++++ Q G +I + +W A GR+ + E+ SS+++ V
Sbjct 15 ELPEARKIMKQIGAQIPRIVESIQVDWSA--LGRV---VAEEV------KSSEDMPPVAA 63
Query 68 ----GVAIKFHEQLGMERLIAMFERFSSYEG-------IFYFLGSILAFSTDPEVHFKYI 116
G A+ H+ +G+++++ + + Y+G + G I+ D + +Y
Sbjct 64 STKDGYAVIAHDGIGLKKMVGVSLAGNIYQGAVEIGKCVRISTGGIIPEGADAVIMREYT 123
Query 117 EAAAKLNHTQEVERVCRE 134
E + ++E E +C++
Sbjct 124 ELVRQDQQSEETEIICKQ 141
> ath:AT2G27800 hypothetical protein
Length=442
Score = 28.9 bits (63), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 128 VERVCREGKYYEPQRVKEFLKQAQL 152
V+ CR+GKY E R+ E L++ QL
Sbjct 401 VDESCRKGKYDEATRLLEMLREKQL 425
> ath:AT5G27300 hypothetical protein
Length=550
Score = 28.5 bits (62), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 128 VERVCREGKYYEPQRVKEFLKQAQL 152
V+ CR+GKY E R+ E L++ QL
Sbjct 295 VDESCRKGKYDEATRLLEMLREKQL 319
> mmu:102182 Prmt10, AI931714; protein arginine methyltransferase
10 (putative)
Length=846
Score = 28.5 bits (62), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 36/70 (51%), Gaps = 9/70 (12%)
Query 2 EAAGLYQRALENYTELADVKRVLLQTGGKISQEWMAQFFGRMSPDICVEILTDMLRSSSQ 61
E +GL Q+ L E A + R LLQ+GGKI +++ F + VE T + S+ Q
Sbjct 666 EPSGLIQQEL---MEKAAISRCLLQSGGKIFPQYVLMF------GMLVESQTLVEESAVQ 716
Query 62 NLQAVVGVAI 71
+ +G+ I
Sbjct 717 GTEHTLGLNI 726
> ath:AT5G06680 SPC98; SPC98 (SPINDLE POLE BODY COMPONENT 98);
tubulin binding
Length=838
Score = 28.5 bits (62), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 46/106 (43%), Gaps = 22/106 (20%)
Query 52 LTDMLRSSSQNLQAVVGVAIKFHE-QLGMERLIAMFE---RFSS-----YEGIFYFLGSI 102
L D+L + + L A+VG ++ + Q E L +FE RF S YEGI
Sbjct 694 LDDLLAAHEKYLNAIVGKSLLGEQSQTIRESLFVLFELILRFRSHADRLYEGIH------ 747
Query 103 LAFSTDPEVHFKYIEAAAKLNHTQEVERVCREGKYYEPQRVKEFLK 148
E+ + E+ + N +QE EG+ QR EFL+
Sbjct 748 -------ELQIRSKESGREKNKSQEPGSWISEGRKGLTQRAGEFLQ 786
Lambda K H
0.323 0.136 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40