bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6419_orf1
Length=87
Score E
Sequences producing significant alignments: (Bits) Value
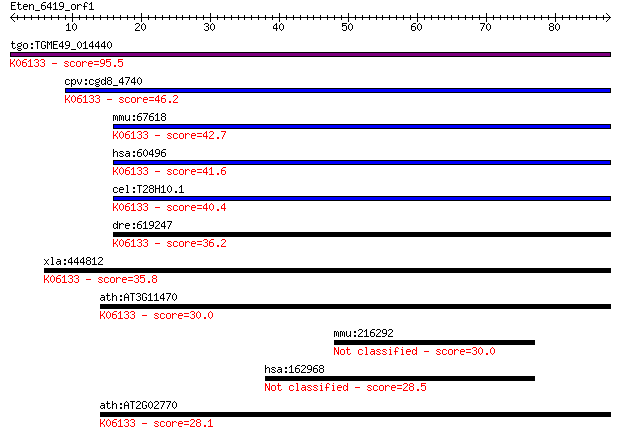
tgo:TGME49_014440 4'-phosphopantetheinyl transferase, putative... 95.5 3e-20
cpv:cgd8_4740 phosphopantetheinyl transferase ; K06133 4'-phos... 46.2 3e-05
mmu:67618 Aasdhppt, 2010309J24Rik, 2810407B07Rik, AASD-PPT, CG... 42.7 3e-04
hsa:60496 AASDHPPT, AASD-PPT, DKFZp566E2346, LYS2, LYS5; amino... 41.6 6e-04
cel:T28H10.1 hypothetical protein; K06133 4'-phosphopantethein... 40.4 0.001
dre:619247 MGC114148; zgc:114148; K06133 4'-phosphopantetheiny... 36.2 0.026
xla:444812 aasdhppt, MGC84206; aminoadipate-semialdehyde dehyd... 35.8 0.037
ath:AT3G11470 4'-phosphopantetheinyl transferase family protei... 30.0 1.6
mmu:216292 cDNA sequence BC067068 30.0 1.8
hsa:162968 ZNF497, FLJ42722, FLJ44773; zinc finger protein 497 28.5
ath:AT2G02770 holo-[acyl-carrier-protein] synthase/ magnesium ... 28.1 6.5
> tgo:TGME49_014440 4'-phosphopantetheinyl transferase, putative
(EC:2.7.8.7); K06133 4'-phosphopantetheinyl transferase [EC:2.7.8.-]
Length=437
Score = 95.5 bits (236), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 47/93 (50%), Positives = 61/93 (65%), Gaps = 6/93 (6%)
Query 1 PIWRPSAGETADFVRFNVSHDEGLVVFGASRHLVGVDCMRCSARG---RDTAEFLQQMKS 57
PIW P A E + F+ FNVSHD GLVV ASR LVGVDCM+ + RG R A+ L+ M S
Sbjct 132 PIWIPHAEERSPFIHFNVSHDTGLVVLSASRLLVGVDCMQVTVRGASSRSVADLLRAMDS 191
Query 58 HCAQKDWAYVMGAH---LKEQQLRRFMRVWTVK 87
C ++ YV+G E++LRRFM++WT+K
Sbjct 192 QCVGRERPYVLGEDSDVSDEEKLRRFMKIWTMK 224
> cpv:cgd8_4740 phosphopantetheinyl transferase ; K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=267
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 42/89 (47%), Gaps = 16/89 (17%)
Query 9 ETADFVRFNVSHDEGLVVFGASRHLVGVDCMRCSARG----------RDTAEFLQQMKSH 58
E+ + FNVSHD +VV S+++VG+D M+ +FL MK+
Sbjct 51 ESDSLLHFNVSHDGDVVVIILSKYMVGIDIMKTELSPSRVQLSNNIMEANEKFLNNMKNV 110
Query 59 CAQKDWAYVMGAHLKEQQLRRFMRVWTVK 87
+W Y+ ++ + +FM WT+K
Sbjct 111 FHPSEWEYI------QKDISKFMHYWTIK 133
> mmu:67618 Aasdhppt, 2010309J24Rik, 2810407B07Rik, AASD-PPT,
CGI-80, LYS2, LYS5; aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl
transferase (EC:2.7.8.-); K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=309
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 36/74 (48%), Gaps = 2/74 (2%)
Query 16 FNVSHDEGLVVFGASRHL-VGVDCMRCSARGRDT-AEFLQQMKSHCAQKDWAYVMGAHLK 73
FN+SH V A + VG+D M+ S GR + EF MK +K+W + + +
Sbjct 107 FNISHQGDYAVLAAEPEVQVGIDIMKTSFPGRGSIPEFFHIMKRKFTKKEWETIRSFNDE 166
Query 74 EQQLRRFMRVWTVK 87
QL F R W +K
Sbjct 167 WTQLDMFYRHWALK 180
> hsa:60496 AASDHPPT, AASD-PPT, DKFZp566E2346, LYS2, LYS5; aminoadipate-semialdehyde
dehydrogenase-phosphopantetheinyl transferase
(EC:2.7.8.-); K06133 4'-phosphopantetheinyl transferase
[EC:2.7.8.-]
Length=309
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 2/74 (2%)
Query 16 FNVSHDEGLVVFGASRHL-VGVDCMRCSARGRDT-AEFLQQMKSHCAQKDWAYVMGAHLK 73
FN+SH V A L VG+D M+ S GR + EF MK K+W + +
Sbjct 107 FNISHQGDYAVLAAEPELQVGIDIMKTSFPGRGSIPEFFHIMKRKFTNKEWETIRSFKDE 166
Query 74 EQQLRRFMRVWTVK 87
QL F R W +K
Sbjct 167 WTQLDMFYRNWALK 180
> cel:T28H10.1 hypothetical protein; K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=299
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 25/74 (33%), Positives = 38/74 (51%), Gaps = 2/74 (2%)
Query 16 FNVSHDEGLVVFGASRHLVGVDCMRCSARGRDTA-EFLQQMKSHCAQKDWAYVMGAHLKE 74
+NVSH LVV +GVD MR + R+TA E + +K H ++ + V G E
Sbjct 97 YNVSHHGDLVVLATGDTRIGVDVMRVNEARRETASEQMNTLKRHFSENEIEMVKGGDKCE 156
Query 75 -QQLRRFMRVWTVK 87
++ F R+W +K
Sbjct 157 LKRWHAFYRIWCLK 170
> dre:619247 MGC114148; zgc:114148; K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=293
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 32/74 (43%), Gaps = 2/74 (2%)
Query 16 FNVSHDEGLVVFGA-SRHLVGVDCMRCSARGRDTA-EFLQQMKSHCAQKDWAYVMGAHLK 73
FNVSH V A + VG+D M+ S G + EF + M +W + A
Sbjct 92 FNVSHQGDYAVLAAEAGRQVGIDVMKTSRPGSSSVQEFFRIMNRQFTDLEWTNIRTAGSD 151
Query 74 EQQLRRFMRVWTVK 87
QL F R W +K
Sbjct 152 WDQLHMFYRHWALK 165
> xla:444812 aasdhppt, MGC84206; aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl
transferase; K06133 4'-phosphopantetheinyl
transferase [EC:2.7.8.-]
Length=302
Score = 35.8 bits (81), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 38/86 (44%), Gaps = 4/86 (4%)
Query 6 SAGETADF--VRFNVSHDEGLVVFGAS-RHLVGVDCMRCSARGRDT-AEFLQQMKSHCAQ 61
+ G ++++ FNVSH V A VGVD M+ G + EF + M +
Sbjct 92 TGGSSSEYPCFNFNVSHQGDYAVLAAEPDRQVGVDIMKTDLPGSSSIEEFFRLMNRQFTE 151
Query 62 KDWAYVMGAHLKEQQLRRFMRVWTVK 87
K+W + + +L F R W +K
Sbjct 152 KEWNSIRSMNNDWARLDMFYRHWALK 177
> ath:AT3G11470 4'-phosphopantetheinyl transferase family protein;
K06133 4'-phosphopantetheinyl transferase [EC:2.7.8.-]
Length=231
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 38/76 (50%), Gaps = 5/76 (6%)
Query 14 VRFNVSHDEGLVVFGASRHL-VGVDCMRCSARGR-DTAEFLQQMKSHCAQKDWAYVMGAH 71
+ FN+SH + L+ G + H+ VG+D + + D F ++ S K ++
Sbjct 55 LHFNISHTDSLIACGVTVHVPVGIDVEDKERKIKHDILAFAERFYSADEVK---FLSTLP 111
Query 72 LKEQQLRRFMRVWTVK 87
E Q + F+++WT+K
Sbjct 112 DPEVQRKEFIKLWTLK 127
> mmu:216292 cDNA sequence BC067068
Length=600
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 48 TAEFLQQMKSHCAQKDWAYVMGAHLKEQQ 76
+ EF Q K CA ++W + M +LKE++
Sbjct 411 SEEFENQHKDRCANENWGFPMCHYLKEER 439
> hsa:162968 ZNF497, FLJ42722, FLJ44773; zinc finger protein 497
Length=498
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 25/46 (54%), Gaps = 7/46 (15%)
Query 38 CMRCSARGRDTAEFLQQMKSH-------CAQKDWAYVMGAHLKEQQ 76
C C R++++ LQ ++H CA+ A+VMG++L E +
Sbjct 304 CAECGKAFRESSQLLQHQRTHTGERPFECAECGQAFVMGSYLAEHR 349
> ath:AT2G02770 holo-[acyl-carrier-protein] synthase/ magnesium
ion binding; K06133 4'-phosphopantetheinyl transferase [EC:2.7.8.-]
Length=661
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 17/75 (22%), Positives = 37/75 (49%), Gaps = 3/75 (4%)
Query 14 VRFNVSHDEGLVVFGASRHL-VGVDCMRCSARGRDTAEFLQQMKSHCAQKDWAYVMGAHL 72
+ FN+SH + L+ G + H+ VG+D + + + L + + + ++
Sbjct 429 LHFNISHTDSLISCGVTVHVPVGIDLEEMERKIKH--DVLALAERFYSADEVKFLSAIPD 486
Query 73 KEQQLRRFMRVWTVK 87
E Q + F+++WT+K
Sbjct 487 PEVQRKEFIKLWTLK 501
Lambda K H
0.327 0.135 0.448
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2026251472
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40