bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6337_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
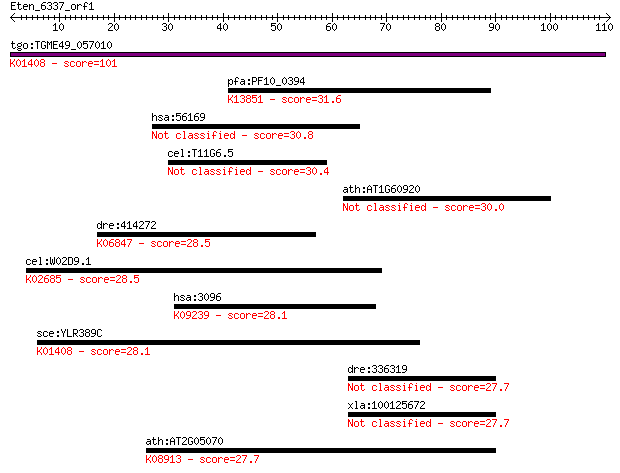
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 101 8e-22
pfa:PF10_0394 rifin; K13851 repetitive interspersed family pro... 31.6 0.58
hsa:56169 GSDMC, MLZE; gasdermin C 30.8 1.1
cel:T11G6.5 hypothetical protein 30.4 1.4
ath:AT1G60920 agl55; agl55 (AGAMOUS-LIKE 55); DNA binding / tr... 30.0 1.9
dre:414272 rgma, MGC110534, MGC192952, dlm, hfe2, id:ibd2030, ... 28.5 5.1
cel:W02D9.1 pri-2; DNA PRImase homolog family member (pri-2); ... 28.5 5.3
hsa:3096 HIVEP1, CIRIP, CRYBP1, MBP-1, PRDII-BF1, Schnurri-1, ... 28.1 6.9
sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1... 28.1 7.2
dre:336319 fj63c11; wu:fj63c11 27.7 9.0
xla:100125672 c17orf28; chromosome 17 open reading frame 28 27.7
ath:AT2G05070 LHCB2.2; chlorophyll binding; K08913 light-harve... 27.7 9.9
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 101 bits (251), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 40/109 (36%), Positives = 69/109 (63%), Gaps = 0/109 (0%)
Query 1 ADPEFSSKVESFETDPYYGVQFRVLDLPQHHAVAMTVLTASPNAFRMPPPLLHIPKASEL 60
ADP+F +SF+ +P+YG+++++ +LP+ + +T SP A+++PP L H+P+ +L
Sbjct 457 ADPQFKRNNDSFDVEPFYGIEYKITNLPKEQRRRLETVTPSPGAYKIPPALKHVPRPEDL 516
Query 61 EILPGLLGLNEPELISEQGGNAGTAVWWQGQGFSALPRVAVQLSGSIAK 109
+LP L G++ PEL+ + + G AVWWQGQG +PRV + +
Sbjct 517 HLLPALGGMSIPELLGDSNTSGGHAVWWQGQGTLPVPRVHANIKARTQR 565
> pfa:PF10_0394 rifin; K13851 repetitive interspersed family protein
Length=309
Score = 31.6 bits (70), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 24/48 (50%), Gaps = 2/48 (4%)
Query 41 SPNAFRMPPPLLHIPKASELEILPGLLGLNEPELISEQGGNAGTAVWW 88
+ +R P L A++ E+ G +G E EL+ Q G GT +W+
Sbjct 186 NKETYRCPQALTTSIYAAKQEVCVGKIG--ETELVCNQLGQGGTPIWF 231
> hsa:56169 GSDMC, MLZE; gasdermin C
Length=508
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 23/46 (50%), Gaps = 8/46 (17%)
Query 27 LPQHHAVAMTVLTASPNAFRMPPPLL--------HIPKASELEILP 64
LP H ++ T+ AS N ++ P L H+PK ++ ILP
Sbjct 259 LPSFHTISPTLFNASSNDMKLKPELFLTQQFLSGHLPKYEQVHILP 304
> cel:T11G6.5 hypothetical protein
Length=1406
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 30 HHAVAMTVLTASPNAFRMPPPLLHIPKAS 58
H A T + SP F PPP+LH P+++
Sbjct 614 HSASISTNIPGSPTQFNRPPPVLHSPRSA 642
> ath:AT1G60920 agl55; agl55 (AGAMOUS-LIKE 55); DNA binding /
transcription factor
Length=191
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 17/38 (44%), Gaps = 0/38 (0%)
Query 62 ILPGLLGLNEPELISEQGGNAGTAVWWQGQGFSALPRV 99
++ LL P L + Q +G WW+ Q F L V
Sbjct 72 VVSSLLHNQHPSLPTNQDNRSGLGFWWEDQAFDRLENV 109
> dre:414272 rgma, MGC110534, MGC192952, dlm, hfe2, id:ibd2030,
rgmc, wu:fb72h09, zgc:110534; RGM domain family, member A;
K06847 RGM domain family
Length=449
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 19/40 (47%), Gaps = 0/40 (0%)
Query 17 YYGVQFRVLDLPQHHAVAMTVLTASPNAFRMPPPLLHIPK 56
Y+ Q + DL H + T P A +PPP+L P+
Sbjct 94 YHSAQHGIEDLMSQHNCSKEGPTTQPRARTVPPPVLSPPQ 133
> cel:W02D9.1 pri-2; DNA PRImase homolog family member (pri-2);
K02685 DNA primase large subunit [EC:2.7.7.-]
Length=503
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 31/73 (42%), Gaps = 12/73 (16%)
Query 4 EFSSKVESFETDPYYGVQFRVLDLPQHHAVAMTVLTASPNAFRMPP--------PLLHIP 55
EF+ K+ S + D YG R + + VA T ++ + R PP P H
Sbjct 352 EFTKKITSDKFDKEYGYNIRYMYGKEGRRVAQTAMSCATIILRNPPSAVDCHGCPFRH-- 409
Query 56 KASELEILPGLLG 68
SE ++L LG
Sbjct 410 --SEKQVLKQKLG 420
> hsa:3096 HIVEP1, CIRIP, CRYBP1, MBP-1, PRDII-BF1, Schnurri-1,
ZAS1, ZNF40, ZNF40A; human immunodeficiency virus type I enhancer
binding protein 1; K09239 human immunodeficiency virus
type I enhancer-binding protein
Length=2718
Score = 28.1 bits (61), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 31 HAVAMTVLTASPNAFRMPPPLLHIPKASELEI-LPGLL 67
+AV + VLTA+P++ P P HIP L I LP L+
Sbjct 2518 NAVGLQVLTANPSSQSSPAPQAHIPGLQILNIALPTLI 2555
> sce:YLR389C STE23; Metalloprotease involved, with homolog Axl1p,
in N-terminal processing of pro-A-factor to the mature
form; member of the insulin-degrading enzyme family (EC:3.4.24.-);
K01408 insulysin [EC:3.4.24.56]
Length=1027
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 17/70 (24%), Positives = 34/70 (48%), Gaps = 1/70 (1%)
Query 6 SSKVESFETDPYYGVQFRVLDLPQHHAVAMTVLTASPNAFRMPPPLLHIPKASELEILPG 65
S +E+ + +YG ++V+D P M +P A +P P + +++ + G
Sbjct 493 SRSLETDSAEKWYGTAYKVVDYPADLIKNMKSPGLNP-ALTLPRPNEFVSTNFKVDKIDG 551
Query 66 LLGLNEPELI 75
+ L+EP L+
Sbjct 552 IKPLDEPVLL 561
> dre:336319 fj63c11; wu:fj63c11
Length=801
Score = 27.7 bits (60), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 63 LPGLLGLNEPELISEQGGNAGTAVWWQ 89
L GLL + P LI + NAGTA+W++
Sbjct 745 LVGLLPVPHPILIRKYQANAGTAMWFR 771
> xla:100125672 c17orf28; chromosome 17 open reading frame 28
Length=792
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 63 LPGLLGLNEPELISEQGGNAGTAVWWQ 89
L GLL + P LI + NAGTA+W++
Sbjct 736 LVGLLPVPHPILIRKYQANAGTAMWFR 762
> ath:AT2G05070 LHCB2.2; chlorophyll binding; K08913 light-harvesting
complex II chlorophyll a/b binding protein 2
Length=265
Score = 27.7 bits (60), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 30/70 (42%), Gaps = 15/70 (21%)
Query 26 DLPQHHAVAMTVLTASPNAFRMPPPLLHIPKASELEILP------GLLGLNEPELISEQG 79
+ P + L+A P F K ELE++ G LG PE++S+ G
Sbjct 72 EYPGDYGWDTAGLSADPETF---------AKNRELEVIHSRWAMLGALGCTFPEILSKNG 122
Query 80 GNAGTAVWWQ 89
G AVW++
Sbjct 123 VKFGEAVWFK 132
Lambda K H
0.316 0.134 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40