bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6305_orf1
Length=130
Score E
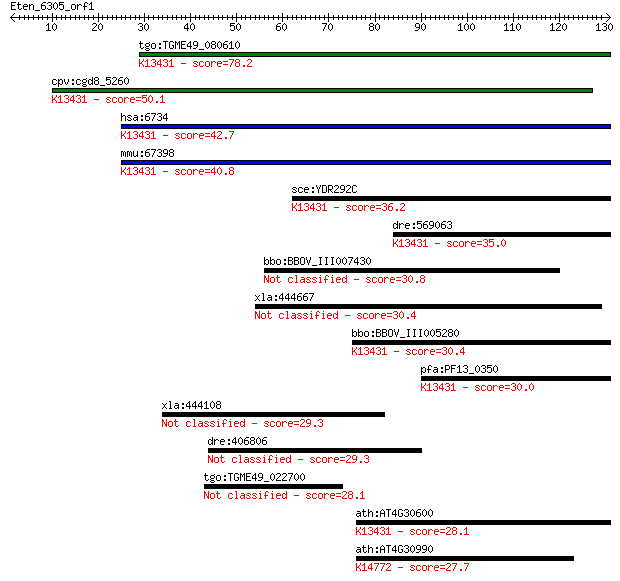
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_080610 signal recognition particle protein, putativ... 78.2 6e-15
cpv:cgd8_5260 Srp101p GTpase. signal recognition particle rece... 50.1 2e-06
hsa:6734 SRPR, DP, MGC17355, MGC3650, MGC9571, Sralpha; signal... 42.7 3e-04
mmu:67398 Srpr, 1300011P19Rik, D11Mgi27; signal recognition pa... 40.8 0.001
sce:YDR292C SRP101; Signal recognition particle (SRP) receptor... 36.2 0.024
dre:569063 srpr, id:ibd1064, wu:fd12f02; signal recognition pa... 35.0 0.053
bbo:BBOV_III007430 17.m07650; 30S ribosomal protein S15 30.8 1.1
xla:444667 samd4b, MGC84256; sterile alpha motif domain contai... 30.4 1.3
bbo:BBOV_III005280 17.m07472; SRP54-type protein, GTPase domai... 30.4 1.4
pfa:PF13_0350 signal recognition particle receptor alpha subun... 30.0 1.6
xla:444108 eapp; E2F-associated phosphoprotein 29.3 3.4
dre:406806 cand1, fb78a05, wu:fb78a05, zgc:55729; cullin-assoc... 29.3 3.4
tgo:TGME49_022700 hypothetical protein 28.1 7.5
ath:AT4G30600 signal recognition particle receptor alpha subun... 28.1 8.0
ath:AT4G30990 binding; K14772 U3 small nucleolar RNA-associate... 27.7 9.9
> tgo:TGME49_080610 signal recognition particle protein, putative
(EC:2.7.4.9); K13431 signal recognition particle receptor
subunit alpha
Length=564
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 51/109 (46%), Positives = 61/109 (55%), Gaps = 7/109 (6%)
Query 29 YSKKTG-----NGDEAAHAIERKTYGADSDSQSSEEETEAAAAAAA--ATAEGSWLSFLP 81
+SKK+ NGD+A +ER TYGAD + SE+E E A A W L
Sbjct 199 FSKKSPHGELPNGDDAVARMERATYGADEALEESEDEEELLEVEMNEEAAAASGWFRSLT 258
Query 82 KSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
LQ GNK+L +DI EVLP FR H + KNVAEEV DLLL SV+ L
Sbjct 259 SKLQSFTGNKLLTNADIAEVLPAFRAHLAEKNVAEEVIDLLLHSVQSRL 307
> cpv:cgd8_5260 Srp101p GTpase. signal recognition particle receptor
alpha subunit ; K13431 signal recognition particle receptor
subunit alpha
Length=548
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/117 (31%), Positives = 54/117 (46%), Gaps = 5/117 (4%)
Query 10 AAGYWTENKVTKKSMEALDYSKKTGNGDEAAHAIERKTYGADSDSQSSEEETEAAAAAAA 69
A+ W KVTKK+ME LDYSK++GN +E+ + Y + D S +E E A
Sbjct 181 ASRQWGNQKVTKKNMEMLDYSKRSGNSEESQSKLS--FYERNEDEVSDDEIEEINQEKKA 238
Query 70 ATAEGSWLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSV 126
+ W +FL N L +E+ L + KNVA E+ ++ SV
Sbjct 239 VSY---WSNFLSSQFGILNNNTTLTAEMVEKPLQKLKSQLVEKNVASEIAQDIIDSV 292
> hsa:6734 SRPR, DP, MGC17355, MGC3650, MGC9571, Sralpha; signal
recognition particle receptor (docking protein); K13431 signal
recognition particle receptor subunit alpha
Length=610
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 51/115 (44%), Gaps = 10/115 (8%)
Query 25 EALDYSKKTGNG------DEAAHAIERKTYGA---DSDSQSSEEETEAAAAAAAATAEGS 75
E LDYS T NG E + I G D D SS++E A + + +G+
Sbjct 229 EVLDYSTPTTNGTPEAALSEDINLIRGTGSGGQLQDLDCSSSDDEGAAQNSTKPSATKGT 288
Query 76 WLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
L + L+ G+K L D+E VL R H AKNVA ++ L SV L
Sbjct 289 -LGGMFGMLKGLVGSKSLSREDMESVLDKMRDHLIAKNVAADIAVQLCESVANKL 342
> mmu:67398 Srpr, 1300011P19Rik, D11Mgi27; signal recognition
particle receptor ('docking protein'); K13431 signal recognition
particle receptor subunit alpha
Length=636
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 39/113 (34%), Positives = 49/113 (43%), Gaps = 7/113 (6%)
Query 25 EALDYSKKTGNGD-EAAHA----IERKTY--GADSDSQSSEEETEAAAAAAAATAEGSWL 77
E LDYS T NG EAA + + R T G D S + E A +A L
Sbjct 256 EVLDYSTPTTNGTPEAALSEDINLIRGTGPGGQLQDLDCSSSDDEGATQNTKPSATKGTL 315
Query 78 SFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
+ L+ G+K L D+E VL R H AKNVA ++ L SV L
Sbjct 316 GGMFGMLKGLVGSKSLSREDMESVLDKMRDHLIAKNVAADIAVQLCESVANKL 368
> sce:YDR292C SRP101; Signal recognition particle (SRP) receptor
- alpha subunit; contain GTPase domains; involved in SRP-dependent
protein targeting; interacts with Srp102p; K13431
signal recognition particle receptor subunit alpha
Length=621
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 34/69 (49%), Gaps = 7/69 (10%)
Query 62 EAAAAAAAATAEGSWLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDL 121
EA + +TA G FL K + GNK ++ESD++ VL Q KNVA E D
Sbjct 282 EAKNSGYVSTAFG----FLQKHV---LGNKTINESDLKSVLEKLTQQLITKNVAPEAADY 334
Query 122 LLLSVKKNL 130
L V +L
Sbjct 335 LTQQVSHDL 343
> dre:569063 srpr, id:ibd1064, wu:fd12f02; signal recognition
particle receptor (docking protein); K13431 signal recognition
particle receptor subunit alpha
Length=650
Score = 35.0 bits (79), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 84 LQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
L+ G+K L + D+E VL + H AKNVA E+ L SV K L
Sbjct 336 LKGLVGSKNLTQEDMEPVLDKMKDHLIAKNVAAEIASQLCDSVAKKL 382
> bbo:BBOV_III007430 17.m07650; 30S ribosomal protein S15
Length=299
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 38/64 (59%), Gaps = 0/64 (0%)
Query 56 SSEEETEAAAAAAAATAEGSWLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVA 115
+SE++T++ ++ T+ S+ SFLPK +K + + DI++ P FR+ + + V
Sbjct 65 TSEDQTQSEDSSKTETSTESFDSFLPKKKKKREPKNPVVKRDIDKFEPLFRKIPALRYVK 124
Query 116 EEVT 119
+EVT
Sbjct 125 KEVT 128
> xla:444667 samd4b, MGC84256; sterile alpha motif domain containing
4B
Length=676
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 36/78 (46%), Gaps = 3/78 (3%)
Query 54 SQSSEEETEAAAAAAAATAEGSWLSFLP---KSLQKTAGNKVLDESDIEEVLPYFRQHFS 110
S SE+ E +A T +GS + +P KSL+ + + EE++ QH
Sbjct 279 SSGSEQTEEVSAGRNTFTEDGSGMKDVPTWLKSLRLHKYASLFAQMSYEEMMTLTEQHLE 338
Query 111 AKNVAEEVTDLLLLSVKK 128
+NV + + LS++K
Sbjct 339 CQNVTKGARHKIALSIQK 356
> bbo:BBOV_III005280 17.m07472; SRP54-type protein, GTPase domain
containing protein; K13431 signal recognition particle receptor
subunit alpha
Length=566
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 75 SWLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
S L + + K AGN VL IEE L + + + KNVA +++ ++ S+ L
Sbjct 254 SVLDTFSQMVLKYAGNMVLTPEVIEEPLKELKLNLNTKNVANDISTMICDSISAGL 309
> pfa:PF13_0350 signal recognition particle receptor alpha subunit,
putative; K13431 signal recognition particle receptor
subunit alpha
Length=576
Score = 30.0 bits (66), Expect = 1.6, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 90 NKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
N ++E DIE++L + +KNVA + D L+ +K+NL
Sbjct 280 NSKIEEEDIEKILHEIKNKLLSKNVAIGICDTLIDRMKENL 320
> xla:444108 eapp; E2F-associated phosphoprotein
Length=293
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 4/51 (7%)
Query 34 GNGDEAAHAIERKTYGADSDSQSSEE---ETEAAAAAAAATAEGSWLSFLP 81
G D+ I R+ +S+S S +E E E+ ++A T EGSW S +P
Sbjct 37 GTPDQKRKMI-RECLTGESESSSDDEFEKEMESELSSAMKTMEGSWQSIVP 86
> dre:406806 cand1, fb78a05, wu:fb78a05, zgc:55729; cullin-associated
and neddylation-dissociated 1
Length=1230
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 44 ERKTYGADSDSQSSEEETEAAAAAAAATAEGSWLSFLPKSLQKTAG 89
E KT D+ S +SEE AA+ A + + G+ +LP LQ+ +G
Sbjct 856 ELKTVILDAFSSASEEVKSAASYALGSISVGNLPEYLPFVLQEISG 901
> tgo:TGME49_022700 hypothetical protein
Length=648
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 43 IERKTYGADSDSQSSEEETEAAAAAAAATA 72
+ERK+ G D DSQ +E++ E+ A TA
Sbjct 244 VERKSDGRDVDSQQAEQQCESPAEPGDTTA 273
> ath:AT4G30600 signal recognition particle receptor alpha subunit
family protein; K13431 signal recognition particle receptor
subunit alpha
Length=634
Score = 28.1 bits (61), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 27/55 (49%), Gaps = 3/55 (5%)
Query 76 WLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLLLLSVKKNL 130
W S + Q G L+ +D+ L ++ KNVAEE+ + L SV+ +L
Sbjct 329 WFSSV---FQSITGKANLERTDLGPALKALKERLMTKNVAEEIAEKLCESVEASL 380
> ath:AT4G30990 binding; K14772 U3 small nucleolar RNA-associated
protein 20
Length=2570
Score = 27.7 bits (60), Expect = 9.9, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 76 WLSFLPKSLQKTAGNKVLDESDIEEVLPYFRQHFSAKNVAEEVTDLL 122
W S L +L+ ++ SD+EE L + + + S + V V D+L
Sbjct 559 WQSLLGTTLRSCYKMTGINHSDLEEALSFAKDYKSCEQVLSPVADVL 605
Lambda K H
0.303 0.119 0.321
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2039374804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40