bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6283_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
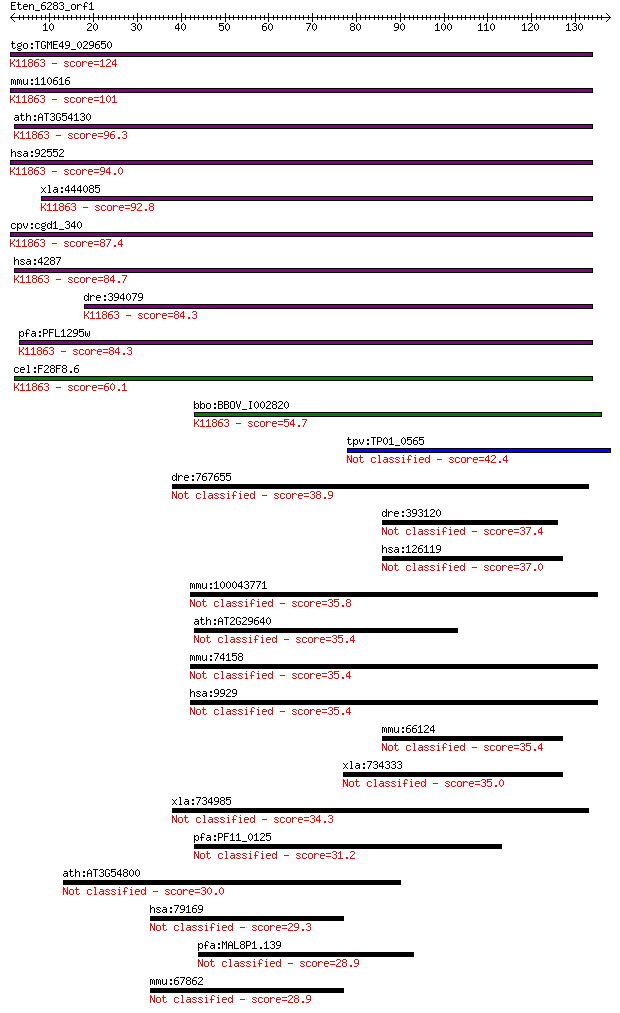
tgo:TGME49_029650 machado-joseph disease protein, putative ; K... 124 7e-29
mmu:110616 Atxn3, 2210008M02Rik, AI463012, AI647473, ATX3, MJD... 101 5e-22
ath:AT3G54130 josephin family protein; K11863 Ataxin-3 [EC:3.4... 96.3 2e-20
hsa:92552 ATXN3L, FLJ59638, MGC168806, MGC168807, MJDL; ataxin... 94.0 1e-19
xla:444085 atxn3, MGC83584; ataxin 3 (EC:3.1.2.15); K11863 Ata... 92.8 3e-19
cpv:cgd1_340 N-terminal machado-Joseph disease protein like do... 87.4 1e-17
hsa:4287 ATXN3, AT3, ATX3, JOS, MJD, MJD1, SCA3; ataxin 3 (EC:... 84.7 7e-17
dre:394079 atxn3, MGC56323, Mjd, zgc:56323; ataxin 3 (EC:3.1.2... 84.3 9e-17
pfa:PFL1295w conserved Plasmodium protein; K11863 Ataxin-3 [EC... 84.3 9e-17
cel:F28F8.6 atx-3; human ATX (ataxin) related family member (a... 60.1 2e-09
bbo:BBOV_I002820 19.m02863; hypothetical protein; K11863 Ataxi... 54.7 8e-08
tpv:TP01_0565 hypothetical protein 42.4 4e-04
dre:767655 josd1, MGC153682, zgc:153682; Josephin domain conta... 38.9 0.005
dre:393120 josd2, MGC55937, zgc:55937; Josephin domain contain... 37.4 0.015
hsa:126119 JOSD2, FLJ29018; Josephin domain containing 2 37.0
mmu:100043771 Gm10651; predicted pseudogene 10651 35.8
ath:AT2G29640 JOSL; JOSL (JOSEPHIN-LIKE PROTEIN) 35.4 0.047
mmu:74158 Josd1, 1300006C06Rik, MGC101975, MGC6306, mKIAA0063;... 35.4 0.049
hsa:9929 JOSD1, KIAA0063, dJ508I15.2; Josephin domain containi... 35.4 0.050
mmu:66124 Josd2, 1110007C05Rik; Josephin domain containing 2 (... 35.4 0.055
xla:734333 josd2, MGC85020; Josephin domain containing 2 35.0
xla:734985 josd1, MGC130880; Josephin domain containing 1 34.3
pfa:PF11_0125 conserved Plasmodium protein 31.2 1.1
ath:AT3G54800 pleckstrin homology (PH) domain-containing prote... 30.0 2.2
hsa:79169 C1orf35, MGC4174, MMTAG2, hMMTAG2; chromosome 1 open... 29.3 4.2
pfa:MAL8P1.139 conserved Plasmodium membrane protein, unknown ... 28.9 4.4
mmu:67862 2310033P09Rik, MGC7354, Mmtag2; RIKEN cDNA 2310033P0... 28.9 5.3
> tgo:TGME49_029650 machado-joseph disease protein, putative ;
K11863 Ataxin-3 [EC:3.4.22.-]
Length=412
Score = 124 bits (312), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 65/139 (46%), Positives = 93/139 (66%), Gaps = 10/139 (7%)
Query 1 AELSSIGLELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIH 60
E+S IG + D++ERQLMAEG + Y+ F E SGNVA DG+FNVSVL+ECLR++ +
Sbjct 41 TEMSKIGYDFDRRERQLMAEGMDPAA-YKEFFDEESGNVAHDGFFNVSVLMECLRKQHVQ 99
Query 61 CICQRNVGEHEEAGV------DFGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFH 114
C+ + + E V + G+ILN +HW A+R+V GTWYNLDS+KP+P +++
Sbjct 100 CL---STSKPEVRAVLADPSREEGFILNLNEHWFAIRKVDGTWYNLDSLKPSPVAMTAEQ 156
Query 115 LEAFLSSLKGQGYTIFIVR 133
L++ L+SL QGY F+ R
Sbjct 157 LKSLLTSLTLQGYVAFVAR 175
> mmu:110616 Atxn3, 2210008M02Rik, AI463012, AI647473, ATX3, MJD1,
Mjd, Sca3, ataxin-3; ataxin 3 (EC:3.4.19.12); K11863 Ataxin-3
[EC:3.4.22.-]
Length=291
Score = 101 bits (252), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 51/137 (37%), Positives = 84/137 (61%), Gaps = 5/137 (3%)
Query 1 AELSSIGLELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIH 60
ELSSI +LD++ER MAEGG S+DY+ FL + SGN+ G+F++ V+ L+ +
Sbjct 31 VELSSIAHQLDEEERLRMAEGGVTSEDYRTFLQQPSGNMDDSGFFSIQVISNALKVWGLE 90
Query 61 CICQRNVGEHEEAGVD----FGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLE 116
I N E++ +D +I N+ +HW +R++ W+NL+S+ P +IS +L
Sbjct 91 LIL-FNSPEYQRLRIDPINERSFICNYKEHWFTVRKLGKQWFNLNSLLTGPELISDTYLA 149
Query 117 AFLSSLKGQGYTIFIVR 133
FL+ L+ +GY+IF+V+
Sbjct 150 LFLAQLQQEGYSIFVVK 166
> ath:AT3G54130 josephin family protein; K11863 Ataxin-3 [EC:3.4.22.-]
Length=280
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 58/140 (41%), Positives = 82/140 (58%), Gaps = 12/140 (8%)
Query 2 ELSSIGLELDQKERQLMAEG----GTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRK 57
+L+++ +LD KERQ+M EG G D FLAE S NV+ G F++ VL + L
Sbjct 38 DLAAVAADLDGKERQVMLEGAAVGGFAPGD---FLAEESHNVSLGGDFSIQVLQKALEVW 94
Query 58 RIHCICQRNVGEHEEAGVD----FGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSF 113
+ I N + E A +D +I + DHW +R+V+G WYN DS+ AP +S F
Sbjct 95 DLQVI-PLNCPDAEPAQIDPELESAFICHLHDHWFCIRKVNGEWYNFDSLLAAPQHLSKF 153
Query 114 HLEAFLSSLKGQGYTIFIVR 133
+L AFL SLKG G++IFIV+
Sbjct 154 YLSAFLDSLKGAGWSIFIVK 173
> hsa:92552 ATXN3L, FLJ59638, MGC168806, MGC168807, MJDL; ataxin
3-like (EC:3.4.19.12); K11863 Ataxin-3 [EC:3.4.22.-]
Length=355
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 49/137 (35%), Positives = 81/137 (59%), Gaps = 5/137 (3%)
Query 1 AELSSIGLELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIH 60
EL+SI +LD++ER MAEGG S++Y AFL + S N+ G+F++ V+ L+ +
Sbjct 31 VELASIAHQLDEEERMRMAEGGVTSEEYLAFLQQPSENMDDTGFFSIQVISNALKFWGLE 90
Query 61 CICQRNVGEHEEAGVD----FGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLE 116
I N E+++ G+D +I N+ HW +R+ W+NL+S+ P +IS L
Sbjct 91 -IIHFNNPEYQKLGIDPINERSFICNYKQHWFTIRKFGKHWFNLNSLLAGPELISDTCLA 149
Query 117 AFLSSLKGQGYTIFIVR 133
FL+ L+ Q Y++F+V+
Sbjct 150 NFLARLQQQAYSVFVVK 166
> xla:444085 atxn3, MGC83584; ataxin 3 (EC:3.1.2.15); K11863 Ataxin-3
[EC:3.4.22.-]
Length=316
Score = 92.8 bits (229), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 44/130 (33%), Positives = 80/130 (61%), Gaps = 5/130 (3%)
Query 8 LELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHCICQRNV 67
++LD++ER MAEGG ++DY+ F+ + SGN+ G+F++ V+ + L + I N
Sbjct 1 MQLDEQERMRMAEGGLATEDYRTFMQQPSGNMDDSGFFSIQVISDALGVWGLELIL-FNS 59
Query 68 GEHEEAGV----DFGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLEAFLSSLK 123
E+ G+ + +I N+ +HW +R++ W+NL+S+ P +IS +L FL+ L+
Sbjct 60 REYRNRGINPINERAFIGNYKEHWFTVRKLGKQWFNLNSLLTGPELISDTYLALFLAQLQ 119
Query 124 GQGYTIFIVR 133
+GY+IF+V+
Sbjct 120 QEGYSIFVVK 129
> cpv:cgd1_340 N-terminal machado-Joseph disease protein like
domain, C-terminal UBX, DNA repair like domain ; K11863 Ataxin-3
[EC:3.4.22.-]
Length=397
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 46/136 (33%), Positives = 73/136 (53%), Gaps = 6/136 (4%)
Query 1 AELSSIGLELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIH 60
A LS I E+D ER+L+ + + ++ S N + DG+F++ VL ECL+R
Sbjct 37 AFLSKIAYEIDDMERRLLEKSNS---TFKTISDNNSQNASYDGFFSIMVLQECLQRHGYS 93
Query 61 CICQRNVGEHEE---AGVDFGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLEA 117
CI N + GYI+N ++HW ++R V G W+NLDS+K AP I F +
Sbjct 94 CIPAANPRVQDYILYPSSCCGYIINSSEHWTSIRCVKGKWFNLDSLKAAPIHIDYFEVSK 153
Query 118 FLSSLKGQGYTIFIVR 133
+L + G ++F+V+
Sbjct 154 YLQEIMFSGKSVFVVQ 169
> hsa:4287 ATXN3, AT3, ATX3, JOS, MJD, MJD1, SCA3; ataxin 3 (EC:3.4.19.12);
K11863 Ataxin-3 [EC:3.4.22.-]
Length=346
Score = 84.7 bits (208), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 47/132 (35%), Positives = 74/132 (56%), Gaps = 12/132 (9%)
Query 2 ELSSIGLELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHC 61
ELSSI +LD++ER MAEGG S+DY+ FL S + G + +R RI
Sbjct 32 ELSSIAHQLDEEERMRMAEGGVTSEDYRTFLQVISNALKVWGLELILFNSPEYQRLRIDP 91
Query 62 ICQRNVGEHEEAGVDFGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLEAFLSS 121
I +R+ +I N+ +HW +R++ W+NL+S+ P +IS +L FL+
Sbjct 92 INERS------------FICNYKEHWFTVRKLGKQWFNLNSLLTGPELISDTYLALFLAQ 139
Query 122 LKGQGYTIFIVR 133
L+ +GY+IF+V+
Sbjct 140 LQQEGYSIFVVK 151
> dre:394079 atxn3, MGC56323, Mjd, zgc:56323; ataxin 3 (EC:3.1.2.15);
K11863 Ataxin-3 [EC:3.4.22.-]
Length=266
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 39/120 (32%), Positives = 72/120 (60%), Gaps = 5/120 (4%)
Query 18 MAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHCICQRNVGEHEEAGVD- 76
MAEGG +++Y+ FL + SGN+ G+F++ V+ L + + N E+++ +D
Sbjct 3 MAEGGVQTEEYRTFLQQPSGNMDDSGFFSIQVISNALGVWGLEIVL-FNSREYQQLQMDP 61
Query 77 ---FGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLEAFLSSLKGQGYTIFIVR 133
+I N+ +HW +R++ W+NL+S+ P +IS +L FL+ L+ +GY+IF++R
Sbjct 62 MHEKAFICNYKEHWFTVRKLGQQWFNLNSLLTGPELISDTYLALFLAQLQQEGYSIFVIR 121
> pfa:PFL1295w conserved Plasmodium protein; K11863 Ataxin-3 [EC:3.4.22.-]
Length=381
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 48/137 (35%), Positives = 74/137 (54%), Gaps = 16/137 (11%)
Query 3 LSSIGLELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHCI 62
L+SIG ELD+KE + + + S NV DG+ N+SV+IE LRR I
Sbjct 36 LASIGKELDEKENEFLRSSSN------DLVRNNSFNVLDDGFINISVIIESLRRMNI--- 86
Query 63 CQRNVGEHEEAGV------DFGYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLE 116
++V E + + D GYI N +HW ++R++H TWY LDS+K +P I +L+
Sbjct 87 LLKHVYEEDLIKIISSNHQDIGYICNLQEHWFSIRKIHNTWYVLDSLKSSPLYIKDMNLK 146
Query 117 AFLSSLKGQGYTIFIVR 133
+ + + + Y IF V+
Sbjct 147 FYFNDV-IKKYHIFSVQ 162
> cel:F28F8.6 atx-3; human ATX (ataxin) related family member
(atx-3); K11863 Ataxin-3 [EC:3.4.22.-]
Length=317
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 64/140 (45%), Gaps = 21/140 (15%)
Query 2 ELSSIGLELDQKERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHC 61
+L + +++D+ E+Q++ S N+ GYF++ VL + L +
Sbjct 38 DLRDLAIQMDKMEQQILGNANPTPG--------RSENMNESGYFSIQVLEKALETFSLKL 89
Query 62 ICQRNVGEHEEAGVDF--------GYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSF 113
N A VD+ YI N +HW LR+ W+ L+S+ P ++S
Sbjct 90 TNIEN-----PAMVDYKNNPLTARAYICNLREHWFVLRKFGNQWFELNSVNRGPKLLSDT 144
Query 114 HLEAFLSSLKGQGYTIFIVR 133
++ FL + +GY+IF+V+
Sbjct 145 YVSMFLHQVSSEGYSIFVVQ 164
> bbo:BBOV_I002820 19.m02863; hypothetical protein; K11863 Ataxin-3
[EC:3.4.22.-]
Length=320
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 6/99 (6%)
Query 43 GYFNVSVLIECLRRKRIHC----ICQRNVGEHEEAGVDFGYILNHADHWLALRRVHGT-W 97
G ++V+VLIE LR + HC + + ++ G+I N +HW +R + W
Sbjct 22 GDYDVTVLIEALRLRNFHCNYYTLNDMKLSTFQQRNDKCGFICNAHEHWFCVRMLMTDRW 81
Query 98 YNLDSMKPAPTVISSFHLEAFLSS-LKGQGYTIFIVRPA 135
+ LDS++P P I L +F+S+ L+ + +F+V P+
Sbjct 82 FILDSLRPGPQEIDYGSLYSFISTLLENKKGAVFLVSPS 120
> tpv:TP01_0565 hypothetical protein
Length=231
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 34/61 (55%), Gaps = 1/61 (1%)
Query 78 GYILNHADHWLALRRVHGTWYNLDSMKPAPTVISSFHLEAFLSS-LKGQGYTIFIVRPAR 136
G+I N +HW ++R++HG W LDS++ P ++ L ++ L+ +F+V P
Sbjct 9 GFICNIQEHWFSIRKLHGEWCILDSLRDGPVLVEYGWLHDYIKEILETSRGVVFLVIPNN 68
Query 137 G 137
G
Sbjct 69 G 69
> dre:767655 josd1, MGC153682, zgc:153682; Josephin domain containing
1
Length=235
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 54/114 (47%), Gaps = 20/114 (17%)
Query 38 NVAADGYFNVSVLIECLRRKRIHCIC---QRNVGEHEEAGVDFGYILN------------ 82
N+ +G ++V+V++ L+ + + +R+VG V G+ILN
Sbjct 108 NMLGNGNYDVNVIMAALQTRGFEAVWWDKRRDVGSIALPNVT-GFILNVPSNLRWGPLRL 166
Query 83 --HADHWLALRRVHGTWYNLDSMKPAPTVI-SSFHLEAFLS-SLKGQGYTIFIV 132
HW+ +R V G +YNLDS +P I ++ L FL L+G+ + +V
Sbjct 167 PLKRQHWIGVREVAGVYYNLDSKLRSPHAIGTADELRKFLRHQLRGKNCELLLV 220
> dre:393120 josd2, MGC55937, zgc:55937; Josephin domain containing
2
Length=184
Score = 37.4 bits (85), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Query 86 HWLALRRVHGTWYNLDSMKPAPTVI-SSFHLEAFLSSLKGQ 125
HWLA+R VHG +YNLDS P I L +FL+ Q
Sbjct 117 HWLAVREVHGHFYNLDSKLKGPACIGGETELRSFLTEQLSQ 157
> hsa:126119 JOSD2, FLJ29018; Josephin domain containing 2
Length=188
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 86 HWLALRRVHGTWYNLDSMKPAPTVISSFH-LEAFLSSLKGQG 126
HW+ALR+V G +YNLDS AP + + AFL++ QG
Sbjct 125 HWVALRQVDGVYYNLDSKLRAPEALGDEDGVRAFLAAALAQG 166
> mmu:100043771 Gm10651; predicted pseudogene 10651
Length=235
Score = 35.8 bits (81), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 31/112 (27%), Positives = 50/112 (44%), Gaps = 20/112 (17%)
Query 42 DGYFNVSVLIECLRRKRIHCIC---QRNVGEHEEAGVDFGYILN--------------HA 84
+G ++V+V++ L+ K + +R+VG V G+I+N
Sbjct 112 NGNYDVNVIMAALQTKGYEAVWWDKRRDVGVIALTNV-MGFIMNLPSSLCWGPLKLPLKR 170
Query 85 DHWLALRRVHGTWYNLDSMKPAPTVI-SSFHLEAFLS-SLKGQGYTIFIVRP 134
HW+ +R V G +YNLDS P I L FL L+G+ + +V P
Sbjct 171 QHWICVREVGGAYYNLDSKLKMPEWIGGESELRKFLKYHLRGKNCELLLVVP 222
> ath:AT2G29640 JOSL; JOSL (JOSEPHIN-LIKE PROTEIN)
Length=360
Score = 35.4 bits (80), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 38/75 (50%), Gaps = 15/75 (20%)
Query 43 GYFNVSVLIECLRRKRIHCIC-QRNVG----EHEEAGVDFGYILN----------HADHW 87
G ++V+V+I L K + + +G + ++A G +LN + HW
Sbjct 71 GNYDVNVMITALEGKGKSVVWHDKRIGASSIDLDDADTLMGIVLNVPVKRYGGLWRSRHW 130
Query 88 LALRRVHGTWYNLDS 102
+ +R+++G WYNLDS
Sbjct 131 VVVRKINGVWYNLDS 145
> mmu:74158 Josd1, 1300006C06Rik, MGC101975, MGC6306, mKIAA0063;
Josephin domain containing 1 (EC:3.4.19.12)
Length=202
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 31/112 (27%), Positives = 50/112 (44%), Gaps = 20/112 (17%)
Query 42 DGYFNVSVLIECLRRKRIHCIC---QRNVGEHEEAGVDFGYILN--------------HA 84
+G ++V+V++ L+ K + +R+VG V G+I+N
Sbjct 79 NGNYDVNVIMAALQTKGYEAVWWDKRRDVGVIALTNV-MGFIMNLPSSLCWGPLKLPLKR 137
Query 85 DHWLALRRVHGTWYNLDSMKPAPTVI-SSFHLEAFLS-SLKGQGYTIFIVRP 134
HW+ +R V G +YNLDS P I L FL L+G+ + +V P
Sbjct 138 QHWICVREVGGAYYNLDSKLKMPEWIGGESELRKFLKYHLRGKNCELLLVVP 189
> hsa:9929 JOSD1, KIAA0063, dJ508I15.2; Josephin domain containing
1
Length=202
Score = 35.4 bits (80), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 31/112 (27%), Positives = 50/112 (44%), Gaps = 20/112 (17%)
Query 42 DGYFNVSVLIECLRRKRIHCIC---QRNVGEHEEAGVDFGYILN--------------HA 84
+G ++V+V++ L+ K + +R+VG V G+I+N
Sbjct 79 NGNYDVNVIMAALQTKGYEAVWWDKRRDVGVIALTNV-MGFIMNLPSSLCWGPLKLPLKR 137
Query 85 DHWLALRRVHGTWYNLDSMKPAPTVI-SSFHLEAFLS-SLKGQGYTIFIVRP 134
HW+ +R V G +YNLDS P I L FL L+G+ + +V P
Sbjct 138 QHWICVREVGGAYYNLDSKLKMPEWIGGESELRKFLKHHLRGKNCELLLVVP 189
> mmu:66124 Josd2, 1110007C05Rik; Josephin domain containing 2
(EC:3.4.19.12)
Length=188
Score = 35.4 bits (80), Expect = 0.055, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 86 HWLALRRVHGTWYNLDSMKPAPTVISSFH-LEAFLSSLKGQG 126
HW+ALR+V G +YNLDS AP + + FL++ QG
Sbjct 125 HWVALRQVDGIYYNLDSKLRAPEALGDEDGVRTFLAAALAQG 166
> xla:734333 josd2, MGC85020; Josephin domain containing 2
Length=184
Score = 35.0 bits (79), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 31/65 (47%), Gaps = 15/65 (23%)
Query 77 FGYILNHAD--------------HWLALRRVHGTWYNLDSMKPAPTVI-SSFHLEAFLSS 121
FG+ILN HW+A+R++ G +YNLDS AP + + L+ FL
Sbjct 93 FGFILNIPSPVSLGFLSLPITRKHWIAVRQIDGVYYNLDSKLKAPIKLGGTKELKEFLHG 152
Query 122 LKGQG 126
+G
Sbjct 153 CMSRG 157
> xla:734985 josd1, MGC130880; Josephin domain containing 1
Length=201
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 52/114 (45%), Gaps = 20/114 (17%)
Query 38 NVAADGYFNVSVLIECLRRKRIHCIC---QRNVGEHEEAGVDFGYILN------------ 82
N+ +G ++V+V++ L+ K + +R+V + V G+I+N
Sbjct 74 NMLGNGNYDVNVIMAALQTKGYEAVWWDKRRDVNLISLSNVT-GFIMNLPSSLSWGPLKL 132
Query 83 --HADHWLALRRVHGTWYNLDSMKPAPTVI-SSFHLEAFLS-SLKGQGYTIFIV 132
HW+ +R V G +YNLDS P I S L FL L+G+ + +V
Sbjct 133 PLKRQHWICVREVGGNYYNLDSKLKRPEWIGSEDDLRKFLRYHLRGKNCELLLV 186
> pfa:PF11_0125 conserved Plasmodium protein
Length=236
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 21/98 (21%), Positives = 42/98 (42%), Gaps = 29/98 (29%)
Query 43 GYFNVSVLIECLRRKRIHC-------ICQRNVGEHEEAGVDFG-----------YILN-- 82
G FN+SVL + + + ICQ+ + +H+ + + F +I+N
Sbjct 106 GNFNISVLYFLMNKHNMELQWVDNKEICQK-LKDHKNSAILFNDEQLNDKTLIAFIINIV 164
Query 83 --------HADHWLALRRVHGTWYNLDSMKPAPTVISS 112
H H+ +R++ +W+ LDS P ++ +
Sbjct 165 KLKFFDIYHHRHFYTIRKISDSWFKLDSSLNKPILLPT 202
> ath:AT3G54800 pleckstrin homology (PH) domain-containing protein
/ lipid-binding START domain-containing protein
Length=733
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 1/77 (1%)
Query 13 KERQLMAEGGTHSKDYQAFLAEGSGNVAADGYFNVSVLIECLRRKRIHCICQRNVGEHEE 72
KE + G H D+ A +A G + ++ FN + + LR + C + NV EH +
Sbjct 193 KEAKDWDSRGRHWDDHPAIMAVGVIDGTSEDIFNTLMSLGPLRSEWDFCFYKGNVVEHLD 252
Query 73 AGVDFGYILNHADHWLA 89
D ++ ++D WL
Sbjct 253 GHTDIIHLQLYSD-WLP 268
> hsa:79169 C1orf35, MGC4174, MMTAG2, hMMTAG2; chromosome 1 open
reading frame 35
Length=263
Score = 29.3 bits (64), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 33 AEGSGNVAADGYFNVSVLIECLRRKRIHCICQRNVGEHEEAGVD 76
AE +AA GY NV L ++ +C+R G+ EE GVD
Sbjct 73 AEREALLAALGYKNVKKQPTGLSKEDFAEVCKREGGDPEEKGVD 116
> pfa:MAL8P1.139 conserved Plasmodium membrane protein, unknown
function
Length=5910
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 15/54 (27%), Positives = 29/54 (53%), Gaps = 10/54 (18%)
Query 44 YFNVSVLIECLRRKRIHCIC-----QRNVGEHEEAGVDFGYILNHADHWLALRR 92
Y N ++++ +RR+ C+C N+GE + + +D N + H+LA R+
Sbjct 1826 YGNKLLILKDVRRREDQCVCVGNYNDDNMGEKQSSEID-----NKSKHYLASRK 1874
> mmu:67862 2310033P09Rik, MGC7354, Mmtag2; RIKEN cDNA 2310033P09
gene
Length=260
Score = 28.9 bits (63), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 33 AEGSGNVAADGYFNVSVLIECLRRKRIHCICQRNVGEHEEAGVD 76
AE +AA GY NV L ++ IC+R G+ EE GVD
Sbjct 73 AEREALLAALGYKNVRKQPTGLSKEDFVEICKREGGDPEEKGVD 116
Lambda K H
0.322 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40