bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6252_orf1
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
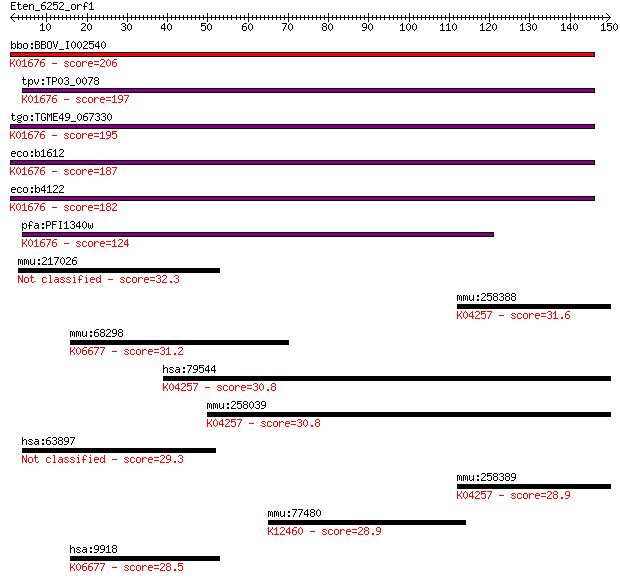
bbo:BBOV_I002540 19.m02245; fumarate hydratase (EC:4.2.1.2); K... 206 3e-53
tpv:TP03_0078 fumarate hydratase (EC:4.2.1.2); K01676 fumarate... 197 1e-50
tgo:TGME49_067330 fumarase, putative (EC:4.2.1.2 4.2.1.32); K0... 195 4e-50
eco:b1612 fumA, ECK1607, JW1604; fumarate hydratase (fumarase ... 187 9e-48
eco:b4122 fumB, ECK4115, JW4083; anaerobic class I fumarate hy... 182 4e-46
pfa:PFI1340w fumarate hydratase, putative (EC:4.2.1.2); K01676... 124 8e-29
mmu:217026 Heatr6, 2700008B19Rik, BB135312, BB249351, FLJ22087... 32.3 0.57
mmu:258388 Olfr1279, MGC130468, MOR245-12; olfactory receptor ... 31.6 0.92
mmu:68298 Ncapd2, 2810406C15Rik, 2810465G24Rik, CAP-D2, CNAP1,... 31.2 1.4
hsa:79544 OR4K1, OR14-19; olfactory receptor, family 4, subfam... 30.8 1.5
mmu:258039 Olfr728, MOR246-1P; olfactory receptor 728; K04257 ... 30.8 1.7
hsa:63897 HEATR6, ABC1, DKFZp686D22141, FLJ22087, MGC148096; H... 29.3 4.4
mmu:258389 Olfr1278, MOR245-11; olfactory receptor 1278; K0425... 28.9 6.3
mmu:77480 Kidins220, 3110039L19Rik, AI194387, AI316525, C33000... 28.9 6.7
hsa:9918 NCAPD2, CAP-D2, CNAP1, KIAA0159, hCAP-D2; non-SMC con... 28.5 9.1
> bbo:BBOV_I002540 19.m02245; fumarate hydratase (EC:4.2.1.2);
K01676 fumarate hydratase, class I [EC:4.2.1.2]
Length=576
Score = 206 bits (523), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 94/145 (64%), Positives = 120/145 (82%), Gaps = 1/145 (0%)
Query 1 SREAFGDLAHLLRPSHLQQLRNILDDKDSSVNDRAVALELLKNACIAAGRVLPGCQDTGT 60
+ +AF ++ H LRP HLQQLRNILDD ++S NDR VA++LLKNAC+A+G+VLPGCQDTGT
Sbjct 89 TEQAFVEIMHYLRPQHLQQLRNILDDHEASNNDRFVAMQLLKNACVASGKVLPGCQDTGT 148
Query 61 AMVVGKKGQFVFTEGADEEALSKGAYLAYTTDNLRYSQMAPLDMFTEQNTKCNLPAQVDI 120
A+V+GK+G +++EG DE A++KG Y AY RYSQMAP+ MF E +TKCNLPAQ++I
Sbjct 149 AIVLGKRGNRIYSEG-DEAAIAKGVYEAYVNRKFRYSQMAPITMFDEVSTKCNLPAQIEI 207
Query 121 AAVDGSSYEFLFIAKGGGSANKTFL 145
+ +GS Y+FLFIAKGGGSANKTFL
Sbjct 208 YSKEGSFYDFLFIAKGGGSANKTFL 232
> tpv:TP03_0078 fumarate hydratase (EC:4.2.1.2); K01676 fumarate
hydratase, class I [EC:4.2.1.2]
Length=621
Score = 197 bits (501), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 89/142 (62%), Positives = 113/142 (79%), Gaps = 1/142 (0%)
Query 4 AFGDLAHLLRPSHLQQLRNILDDKDSSVNDRAVALELLKNACIAAGRVLPGCQDTGTAMV 63
AF ++ H LR HL QLR + DD +SS NDR V+++LLKNACI+AG+VLPGCQDTGTA+V
Sbjct 136 AFSEIMHFLRTEHLAQLRKVFDDPESSDNDRFVSMQLLKNACISAGKVLPGCQDTGTAIV 195
Query 64 VGKKGQFVFTEGADEEALSKGAYLAYTTDNLRYSQMAPLDMFTEQNTKCNLPAQVDIAAV 123
+GK+G F+F+E D +++KG Y +Y N RYSQM+ L M+ E NTKCNLPAQV++ +
Sbjct 196 IGKRGNFIFSEN-DANSITKGVYDSYMNKNFRYSQMSALSMYEEVNTKCNLPAQVELYSQ 254
Query 124 DGSSYEFLFIAKGGGSANKTFL 145
DG+ YEFLFIAKGGGSANKTFL
Sbjct 255 DGTDYEFLFIAKGGGSANKTFL 276
> tgo:TGME49_067330 fumarase, putative (EC:4.2.1.2 4.2.1.32);
K01676 fumarate hydratase, class I [EC:4.2.1.2]
Length=641
Score = 195 bits (496), Expect = 4e-50, Method: Compositional matrix adjust.
Identities = 94/145 (64%), Positives = 116/145 (80%), Gaps = 0/145 (0%)
Query 1 SREAFGDLAHLLRPSHLQQLRNILDDKDSSVNDRAVALELLKNACIAAGRVLPGCQDTGT 60
++ AF +++ LR SHL+ LR I DD D+S NDR VAL LLKNA IAAGRV PGCQDTGT
Sbjct 146 TKYAFEEISFFLRRSHLESLRRIYDDPDASDNDRFVALTLLKNANIAAGRVFPGCQDTGT 205
Query 61 AMVVGKKGQFVFTEGADEEALSKGAYLAYTTDNLRYSQMAPLDMFTEQNTKCNLPAQVDI 120
A+V+GKKG V T+G+DEEA+SKG + AYT +NLRYSQ+AP DMFTE NT+ NLPA V +
Sbjct 206 AIVLGKKGASVLTDGSDEEAISKGVFEAYTQNNLRYSQVAPRDMFTEYNTRSNLPADVTL 265
Query 121 AAVDGSSYEFLFIAKGGGSANKTFL 145
+ G+++E LF+AKGGGSANKTFL
Sbjct 266 LSTSGNAFELLFVAKGGGSANKTFL 290
> eco:b1612 fumA, ECK1607, JW1604; fumarate hydratase (fumarase
A), aerobic class I (EC:4.2.1.2); K01676 fumarate hydratase,
class I [EC:4.2.1.2]
Length=548
Score = 187 bits (476), Expect = 9e-48, Method: Compositional matrix adjust.
Identities = 90/145 (62%), Positives = 113/145 (77%), Gaps = 0/145 (0%)
Query 1 SREAFGDLAHLLRPSHLQQLRNILDDKDSSVNDRAVALELLKNACIAAGRVLPGCQDTGT 60
+R+AF D + +LRP+H QQ+ +IL D ++S ND+ VAL+ L+N+ IAA VLP CQDTGT
Sbjct 51 ARQAFHDASFMLRPAHQQQVADILRDPEASENDKYVALQFLRNSDIAAKGVLPTCQDTGT 110
Query 61 AMVVGKKGQFVFTEGADEEALSKGAYLAYTTDNLRYSQMAPLDMFTEQNTKCNLPAQVDI 120
A++VGKKGQ V+T G DE AL++G Y Y DNLRYSQ APLDM+ E NT NLPAQ+D+
Sbjct 111 AIIVGKKGQRVWTGGGDEAALARGVYNTYIEDNLRYSQNAPLDMYKEVNTGTNLPAQIDL 170
Query 121 AAVDGSSYEFLFIAKGGGSANKTFL 145
AVDG Y+FL IAKGGGSANKT+L
Sbjct 171 YAVDGDEYKFLCIAKGGGSANKTYL 195
> eco:b4122 fumB, ECK4115, JW4083; anaerobic class I fumarate
hydratase (fumarase B) (EC:4.2.1.2); K01676 fumarate hydratase,
class I [EC:4.2.1.2]
Length=548
Score = 182 bits (461), Expect = 4e-46, Method: Compositional matrix adjust.
Identities = 88/145 (60%), Positives = 111/145 (76%), Gaps = 0/145 (0%)
Query 1 SREAFGDLAHLLRPSHLQQLRNILDDKDSSVNDRAVALELLKNACIAAGRVLPGCQDTGT 60
+++AF D + +LRP+H +Q+ IL D ++S ND+ VAL+ L+N+ IAA VLP CQDTGT
Sbjct 51 AQQAFHDASFMLRPAHQKQVAAILHDPEASENDKYVALQFLRNSEIAAKGVLPTCQDTGT 110
Query 61 AMVVGKKGQFVFTEGADEEALSKGAYLAYTTDNLRYSQMAPLDMFTEQNTKCNLPAQVDI 120
A++VGKKGQ V+T G DEE LSKG Y Y DNLRYSQ A LDM+ E NT NLPAQ+D+
Sbjct 111 AIIVGKKGQRVWTGGGDEETLSKGVYNTYIEDNLRYSQNAALDMYKEVNTGTNLPAQIDL 170
Query 121 AAVDGSSYEFLFIAKGGGSANKTFL 145
AVDG Y+FL +AKGGGSANKT+L
Sbjct 171 YAVDGDEYKFLCVAKGGGSANKTYL 195
> pfa:PFI1340w fumarate hydratase, putative (EC:4.2.1.2); K01676
fumarate hydratase, class I [EC:4.2.1.2]
Length=681
Score = 124 bits (312), Expect = 8e-29, Method: Composition-based stats.
Identities = 59/117 (50%), Positives = 85/117 (72%), Gaps = 1/117 (0%)
Query 4 AFGDLAHLLRPSHLQQLRNILDDKDSSVNDRAVALELLKNACIAAGRVLPGCQDTGTAMV 63
AF ++ L HL+QL++IL DK+SS ND+ VA+ L+KNA I++ + LPGCQDTGTA++
Sbjct 137 AFKEILFFLNKKHLKQLQHILQDKESSKNDKYVAMTLIKNAIISSEQKLPGCQDTGTAII 196
Query 64 VGKKGQFVFTEGADEEALSKGAYLAYTTDNLRYSQMAPLDMFTEQNTKCNLPAQVDI 120
+GKK + + T + + L+ G Y AY +N RYSQ++PL+MF E NTK NLP Q++I
Sbjct 197 LGKKDEDILTT-YEHKYLTLGVYNAYKYNNFRYSQLSPLNMFNEVNTKNNLPCQIEI 252
> mmu:217026 Heatr6, 2700008B19Rik, BB135312, BB249351, FLJ22087;
HEAT repeat containing 6
Length=1184
Score = 32.3 bits (72), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 31/55 (56%), Gaps = 5/55 (9%)
Query 3 EAFGDLAHLLRPSHLQQLR--NILDDKDSSVNDRAV---ALELLKNACIAAGRVL 52
A G+L H L+PSH+++ R I+++ ++ V A+++ NAC A G V
Sbjct 933 RALGNLLHFLQPSHVERPRFAEIIEESIQALISTVVNEAAMKVRWNACYAMGNVF 987
> mmu:258388 Olfr1279, MGC130468, MOR245-12; olfactory receptor
1279; K04257 olfactory receptor
Length=313
Score = 31.6 bits (70), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 112 CNLPAQVDIAAVDGSSYEFLFIAKGGGSANKTFLALLL 149
C+ P + +A DG +EF+ A G + TF LLL
Sbjct 181 CDFPRIIQLACTDGDKFEFVVAANSGFMSMGTFFLLLL 218
> mmu:68298 Ncapd2, 2810406C15Rik, 2810465G24Rik, CAP-D2, CNAP1,
KIAA0159, MGC39046, mKIAA0159; non-SMC condensin I complex,
subunit D2; K06677 condensin complex subunit 1
Length=1392
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 34/59 (57%), Gaps = 5/59 (8%)
Query 16 HLQQLRNILDDKDSSVNDRAVALELLKNACIAAGRVLPGCQD--TGTAMV---VGKKGQ 69
H Q+L +ILDD S +DR+ L LK C A R+L ++ + T+++ +G KG+
Sbjct 92 HSQELSSILDDAALSGSDRSAHLNALKMNCYALIRLLESFENMTSQTSLIDLDIGGKGK 150
> hsa:79544 OR4K1, OR14-19; olfactory receptor, family 4, subfamily
K, member 1; K04257 olfactory receptor
Length=311
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 24/112 (21%), Positives = 51/112 (45%), Gaps = 7/112 (6%)
Query 39 ELLKNACIAAGRVLPGCQDTGTAMVVGKKGQFVFTEGADEE-ALSKGAYLAYTTDNLRYS 97
E++ +A R + C+ + ++ ++ +F + L ++LA+T D L +
Sbjct 111 EMMLLVAMAYDRFIAICKPLHYSTIMNRRLCVIFVSISWAVGVLHSVSHLAFTVD-LPFC 169
Query 98 QMAPLDMFTEQNTKCNLPAQVDIAAVDGSSYEFLFIAKGGGSANKTFLALLL 149
+D F C+LP +++A +D E + + G + FLAL++
Sbjct 170 GPNEVDSFF-----CDLPLVIELACMDTYEMEIMTLTNSGLISLSCFLALII 216
> mmu:258039 Olfr728, MOR246-1P; olfactory receptor 728; K04257
olfactory receptor
Length=311
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 46/101 (45%), Gaps = 7/101 (6%)
Query 50 RVLPGCQDTGTAMVVGKKGQFVFTEGADEE-ALSKGAYLAYTTDNLRYSQMAPLDMFTEQ 108
R + C+ +++ ++ +F + L ++LA+T NL + +D F
Sbjct 122 RFVAICKPLHYNLIMNRRVCIIFVSISWAVGILHSVSHLAFTV-NLPFCGPNEVDSFF-- 178
Query 109 NTKCNLPAQVDIAAVDGSSYEFLFIAKGGGSANKTFLALLL 149
C+LP + +A +D E L +A G + FLAL++
Sbjct 179 ---CDLPLVIKLACMDTYRMEILTLANSGMISLSCFLALII 216
> hsa:63897 HEATR6, ABC1, DKFZp686D22141, FLJ22087, MGC148096;
HEAT repeat containing 6
Length=1181
Score = 29.3 bits (64), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 30/53 (56%), Gaps = 5/53 (9%)
Query 4 AFGDLAHLLRPSHLQQ--LRNILDDKDSSVNDRAV---ALELLKNACIAAGRV 51
A G+L H L+PSH+++ I+++ ++ + A+++ NAC A G V
Sbjct 931 ALGNLLHFLQPSHIEKPTFAEIIEESIQALISTVLTEAAMKVRWNACYAMGNV 983
> mmu:258389 Olfr1278, MOR245-11; olfactory receptor 1278; K04257
olfactory receptor
Length=313
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 112 CNLPAQVDIAAVDGSSYEFLFIAKGGGSANKTFLALLL 149
C+LP + +A +D +F+ A G + TFL L++
Sbjct 179 CDLPQFIKLACIDPKKMQFMVTANSGFISMGTFLLLII 216
> mmu:77480 Kidins220, 3110039L19Rik, AI194387, AI316525, C330002I19Rik,
mKIAA1250; kinase D-interacting substrate 220; K12460
ankyrin repeat-rich membrane spanning protein
Length=1793
Score = 28.9 bits (63), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 65 GKKGQFVFTEGADEEALSKGAYLAYTTDNLRYSQMAPLDMFTEQNTKCN 113
GK+ + EG + +G L Y++ + ++ +PLD TE++ K +
Sbjct 1459 GKENELKQEEGRKSFLMKRGDVLDYSSSGVSTNEASPLDPITEEDEKSD 1507
> hsa:9918 NCAPD2, CAP-D2, CNAP1, KIAA0159, hCAP-D2; non-SMC condensin
I complex, subunit D2; K06677 condensin complex subunit
1
Length=1401
Score = 28.5 bits (62), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 16 HLQQLRNILDDKDSSVNDRAVALELLKNACIAAGRVL 52
H Q+L ILDD S +DR L LK C A R+L
Sbjct 92 HSQELPAILDDTTLSGSDRNAHLNALKMNCYALIRLL 128
Lambda K H
0.318 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3068761412
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40