bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6192_orf3
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
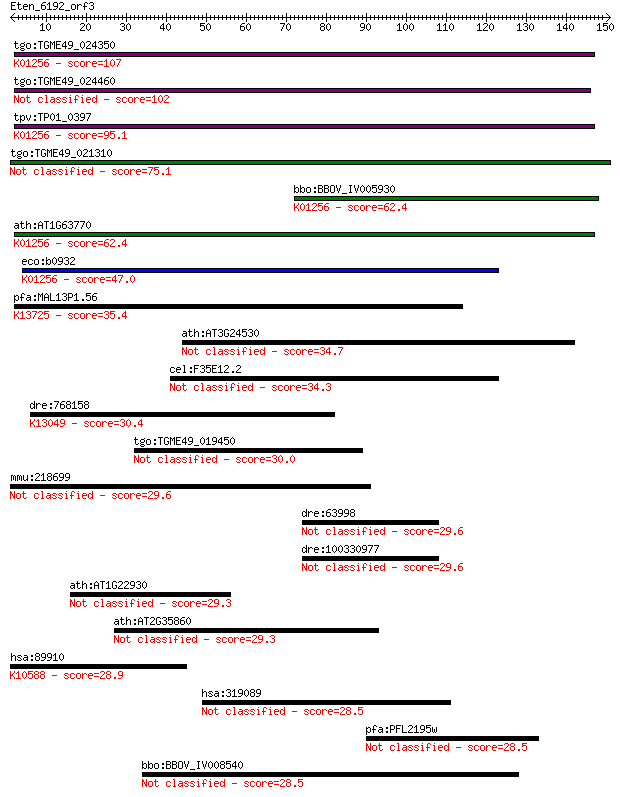
tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2); K0... 107 1e-23
tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2) 102 5e-22
tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2); ... 95.1 6e-20
tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2) 75.1 8e-14
bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01... 62.4 5e-10
ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptida... 62.4 6e-10
eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2... 47.0 2e-05
pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725 ... 35.4 0.071
ath:AT3G24530 AAA-type ATPase family protein / ankyrin repeat ... 34.7 0.13
cel:F35E12.2 hypothetical protein 34.3 0.15
dre:768158 pm20d1.2, pm20d1, zgc:154035; peptidase M20 domain ... 30.4 2.1
tgo:TGME49_019450 WD-repeat protein, putative (EC:3.1.3.2 2.7.... 30.0 3.3
mmu:218699 Pxk, C230080L11Rik, D14Ertd813e, MONaKA; PX domain ... 29.6 3.9
dre:63998 stard3, fc66d12, mln64, mln64-pen, wu:fc66d12; START... 29.6 4.1
dre:100330977 stAR-related lipid transfer protein 3-like 29.6 4.1
ath:AT1G22930 T-complex protein 11 29.3
ath:AT2G35860 FLA16; FLA16 (FASCICLIN-LIKE ARABINOGALACTAN PRO... 29.3 4.8
hsa:89910 UBE3B, DKFZp586K2123, DKFZp686A1051, FLJ45294, MGC13... 28.9 6.9
hsa:319089 TTC6, C14orf25, MGC119358, MGC119360, MGC119361; te... 28.5 7.4
pfa:PFL2195w clathrin coat assembly protein AP180, putative 28.5 8.5
bbo:BBOV_IV008540 23.m06569; helicase 28.5 8.5
> tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1419
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 57/146 (39%), Positives = 88/146 (60%), Gaps = 1/146 (0%)
Query 2 YIDALRTTLTDNTLDNALKALLLQLPDWSTLSTHMKTIDPEALHLAIRSVKADVAAALKT 61
Y++A + TL + D +++A L+LPD ++ M+ IDP AL A SV+ +V LK+
Sbjct 1134 YVEAFKQTLLEQGRDRSIQAYTLRLPDRDGVAQEMEPIDPLALKEATESVRREVGQLLKS 1193
Query 62 EMAKEYEHLTLPAGEEDDMEEES-TGRRKLRNTLLFFLSESRDREAVDRAVKHFTDAKCM 120
++ K Y L+ P E ++ ++S RR+LRN +L+FL+ RD+EA A+ HF AK M
Sbjct 1194 DLLKVYASLSAPESEAEESRDQSEVSRRRLRNVILYFLTGERDKEAAALAMNHFKSAKGM 1253
Query 121 TDRYAGLITLGNIPTAERETAFARFY 146
T++YA L L +I ER A +FY
Sbjct 1254 TEKYAALSILCDIEGPERTAALEQFY 1279
> tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2)
Length=970
Score = 102 bits (253), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 60/147 (40%), Positives = 87/147 (59%), Gaps = 3/147 (2%)
Query 2 YIDALRTTLT--DNTLDNALKALLLQLPDWSTLSTHMKTIDPEALHLAIRSVKADVAAAL 59
Y+ +R + ++++ +K+LLL LP + L + +IDP+A++ A+ SV+ D+ AL
Sbjct 685 YVQTVRDIVIAPESSMGKDIKSLLLSLPTKAQLELAVDSIDPDAINAALASVRRDIVDAL 744
Query 60 KTEMAKEYEHLTLPAG-EEDDMEEESTGRRKLRNTLLFFLSESRDREAVDRAVKHFTDAK 118
EM + Y LTLPAG EE + E GRR LRN LL FL+ S D+++ A HF A
Sbjct 745 GEEMLQLYTELTLPAGTEESGADIEHWGRRALRNELLRFLTASFDQKSAKLASAHFDRAM 804
Query 119 CMTDRYAGLITLGNIPTAERETAFARF 145
M+D+ A L L IP ER+ AF RF
Sbjct 805 VMSDKVAALTVLTEIPGQERDEAFERF 831
> tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1020
Score = 95.1 bits (235), Expect = 6e-20, Method: Composition-based stats.
Identities = 48/145 (33%), Positives = 82/145 (56%), Gaps = 1/145 (0%)
Query 2 YIDALRTTLTDNTLDNALKALLLQLPDWSTLSTHMKTIDPEALHLAIRSVKADVAAALKT 61
Y++A + L + +D+ K L + +PD L++ MK DP L ++R +K ++ +
Sbjct 743 YLEAYKKLLNSD-MDHNEKGLCMSMPDVDILASKMKPYDPGLLFASLRKLKQELGRTFRP 801
Query 62 EMAKEYEHLTLPAGEEDDMEEESTGRRKLRNTLLFFLSESRDREAVDRAVKHFTDAKCMT 121
+ Y+ LTL G++D++ +E RR LRNT+ FL RD E+V+ A+KH+ DAK M
Sbjct 802 TFTEMYKSLTLREGQKDELTKEDMARRFLRNTVFSFLVSMRDMESVELALKHYRDAKVMN 861
Query 122 DRYAGLITLGNIPTAERETAFARFY 146
D+Y + L ++ +R+ FY
Sbjct 862 DKYTAFVQLMHMEFQDRQKVVDDFY 886
> tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2)
Length=966
Score = 75.1 bits (183), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 57/162 (35%), Positives = 81/162 (50%), Gaps = 14/162 (8%)
Query 1 AYIDALRTTLTDNTLDNALKALLLQLPDWSTLSTHM-KTIDPEALHLAIRSVKADVAAAL 59
A+ L L D L A ALLL LP +S L + +DP+A+ A RS+ D+
Sbjct 649 AFQQGLHFALRDMPLSPAFTALLLTLPSYSRLEQDAPRPLDPDAIISARRSLLRDIYYFH 708
Query 60 KTEMAKEYEHLTLPAGEEDDMEE----------ESTGRRKLRNTLLFFLSESRDREAVDR 109
+ + + Y T+P + DD E E RR LR+ LL +++ +RD +
Sbjct 709 RNALDEAYVATTIP--KVDDRERDRQLESAEDPEQWQRRALRSILLEYVTANRDERSAKL 766
Query 110 AVKHFTDAKCMTDRYAGLITLGNIP-TAERETAFARFYEATR 150
A+KHF DA+ MTD+ A L L ++P ERE A FYE R
Sbjct 767 ALKHFKDARVMTDKIAALHVLVDLPFNKEREEALHLFYEEAR 808
> bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01256
aminopeptidase N [EC:3.4.11.2]
Length=846
Score = 62.4 bits (150), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 32/76 (42%), Positives = 45/76 (59%), Gaps = 0/76 (0%)
Query 72 LPAGEEDDMEEESTGRRKLRNTLLFFLSESRDREAVDRAVKHFTDAKCMTDRYAGLITLG 131
L E+D +E++ RR LRNTLL +L D AV+ A+ H+ A+CMTDRY + L
Sbjct 632 LDNNEKDTLEKDDMARRYLRNTLLGYLVCRSDASAVELALGHYRAARCMTDRYYAFVQLM 691
Query 132 NIPTAERETAFARFYE 147
N+ A ++ A FYE
Sbjct 692 NMDFAGKDDVIADFYE 707
> ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptidase
N [EC:3.4.11.2]
Length=987
Score = 62.4 bits (150), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 41/145 (28%), Positives = 71/145 (48%), Gaps = 2/145 (1%)
Query 2 YIDALRTTLTDNTLDNALKALLLQLPDWSTLSTHMKTIDPEALHLAIRSVKADVAAALKT 61
++ L + L+D++LD A + LP + M DP+A+H + V+ +A+ LK
Sbjct 711 FVQGLGSVLSDSSLDKEFIAKAITLPGEGEIMDMMAVADPDAVHAVRKFVRKQLASELKE 770
Query 62 EMAKEYEHLTLPAGEEDDMEEESTGRRKLRNTLLFFLSESRDREAVDRAVKHFTDAKCMT 121
E+ K E+ + E + + RR L+NT L +L+ D ++ A+ + A +T
Sbjct 771 ELLKIVENNR--STEAYVFDHSNMARRALKNTALAYLASLEDPAYMELALNEYKMATNLT 828
Query 122 DRYAGLITLGNIPTAERETAFARFY 146
D++A L L P R+ A FY
Sbjct 829 DQFAALAALSQNPGKTRDDILADFY 853
> eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=870
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/120 (33%), Positives = 54/120 (45%), Gaps = 5/120 (4%)
Query 4 DALRTTLTDNTLDNALKALLLQLPDWSTLSTHMKTIDPEALHLAIRSVKADVAAALKTEM 63
DA R L D +D AL A +L LP + ++ IDP +AI V+ + L TE+
Sbjct 597 DAFRAVLLDEKIDPALAAEILTLPSVNEMAELFDIIDP----IAIAEVREALTRTLATEL 652
Query 64 AKEYEHL-TLPAGEEDDMEEESTGRRKLRNTLLFFLSESRDREAVDRAVKHFTDAKCMTD 122
A E + E +E E +R LRN L FL+ A K F +A MTD
Sbjct 653 ADELLAIYNANYQSEYRVEHEDIAKRTLRNACLRFLAFGETHLADVLVSKQFHEANNMTD 712
> pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725
M1-family aminopeptidase [EC:3.4.11.-]
Length=1085
Score = 35.4 bits (80), Expect = 0.071, Method: Composition-based stats.
Identities = 26/118 (22%), Positives = 51/118 (43%), Gaps = 6/118 (5%)
Query 2 YIDALRTTLTDNTLDNALKALLLQLPDWSTLSTHMKTIDPEALHLAIRSVKADVAAALKT 61
+IDA++ L D D K+ ++ LP + + +D + L + + L
Sbjct 803 FIDAIKYLLEDPHADAGFKSYIVSLPQDRYIINFVSNLDTDVLADTKEYIYKQIGDKLND 862
Query 62 EMAKEYEHLTLPA------GEEDDMEEESTGRRKLRNTLLFFLSESRDREAVDRAVKH 113
K ++ L A +E ++ + R LRNTLL LS+++ ++ ++H
Sbjct 863 VYYKMFKSLEAKADDLTYFNDESHVDFDQMNMRTLRNTLLSLLSKAQYPNILNEIIEH 920
> ath:AT3G24530 AAA-type ATPase family protein / ankyrin repeat
family protein
Length=481
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 51/110 (46%), Gaps = 21/110 (19%)
Query 44 LHLAI-RSVKADVAAALKTEM-------AKEYEHLT----LPAGEEDDMEEESTGRRKLR 91
LHLA+ S+ A + +KT + AK+ E +T LP G+ G KLR
Sbjct 124 LHLAVWYSITAKEISTVKTLLDHNADCSAKDNEGMTPLDHLPQGQ---------GSEKLR 174
Query 92 NTLLFFLSESRDREAVDRAVKHFTDAKCMTDRYAGLITLGNIPTAERETA 141
L +FL E R R A+++ K + + D + ++ L + T R+ A
Sbjct 175 ELLRWFLQEQRKRSALEQCGKTKAKMELLEDELSNIVGLSELKTQLRKWA 224
> cel:F35E12.2 hypothetical protein
Length=428
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 42/84 (50%), Gaps = 9/84 (10%)
Query 41 PEA-LHLAIRSVKADVAAALK-TEMAKEYEHLTLPAGEEDDMEEESTGRRKLRNTLLFFL 98
P A LH+ S + + A + +AK+Y+H+ +P GE + ++N F
Sbjct 109 PSAILHINTISNTSQMFAIYEFVNIAKKYQHVLIPTGEYFSLGN-------MKNRYYTFH 161
Query 99 SESRDREAVDRAVKHFTDAKCMTD 122
S+ +DR A++ AVK+ D + D
Sbjct 162 SKDKDRVALNIAVKNTQDYDALLD 185
> dre:768158 pm20d1.2, pm20d1, zgc:154035; peptidase M20 domain
containing 1, tandem duplicate 2 (EC:3.4.17.-); K13049 carboxypeptidase
PM20D1 [EC:3.4.17.-]
Length=522
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 37/78 (47%), Gaps = 4/78 (5%)
Query 6 LRTTLTDNTLDNALKALLLQLPDWSTLSTHMKTIDPEALHLAIRSVKADV--AAALKTEM 63
LRT N L N A+ ++P S TH+ T + +RSV V ++ ++ E+
Sbjct 60 LRTQQRQNLLANFKAAI--RIPTVSFTETHVNTSALQEFDGLLRSVFPKVFSSSLVRHEV 117
Query 64 AKEYEHLTLPAGEEDDME 81
Y HL AG + D+E
Sbjct 118 VGNYSHLFTIAGTDADLE 135
> tgo:TGME49_019450 WD-repeat protein, putative (EC:3.1.3.2 2.7.11.7)
Length=2650
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Query 32 LSTHMKTIDPEALHLAIRSVKADVAAALKTEMAKEYEHLTLPAGEEDDMEEESTGRR 88
L TH++ + L R + + L++ + E HL LPAGEE + E ++ RR
Sbjct 2535 LETHLRV---KELARQQRLEEQQASVQLESSLQLEVPHLLLPAGEESNDEGDTRRRR 2588
> mmu:218699 Pxk, C230080L11Rik, D14Ertd813e, MONaKA; PX domain
containing serine/threonine kinase
Length=582
Score = 29.6 bits (65), Expect = 3.9, Method: Composition-based stats.
Identities = 26/96 (27%), Positives = 41/96 (42%), Gaps = 6/96 (6%)
Query 1 AYIDALRTTLTDNTLDNALKAL--LLQLPDWS----TLSTHMKTIDPEALHLAIRSVKAD 54
A + L +TL+ N + + LLQ+P +S T S + P L A+R K
Sbjct 366 AVVAVLESTLSCEACKNGMPTVSRLLQMPLFSDVLLTTSEKPQFKIPTKLKEALRIAKEC 425
Query 55 VAAALKTEMAKEYEHLTLPAGEEDDMEEESTGRRKL 90
+ L E + ++H L + EE RRK+
Sbjct 426 IEKRLTEEQKQIHQHRRLTRAQSHHGSEEERKRRKI 461
> dre:63998 stard3, fc66d12, mln64, mln64-pen, wu:fc66d12; START
domain containing 3
Length=448
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 74 AGEEDDMEEESTGRRKLRNTLLFFLSESRDREAV 107
AG EDD++EE GRR + F+ + R+ AV
Sbjct 217 AGSEDDLDEEGLGRRAVTEQEKAFVRQGREAMAV 250
> dre:100330977 stAR-related lipid transfer protein 3-like
Length=448
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 74 AGEEDDMEEESTGRRKLRNTLLFFLSESRDREAV 107
AG EDD++EE GRR + F+ + R+ AV
Sbjct 217 AGSEDDLDEEGLGRRAVTEQEKAFVRQGREAMAV 250
> ath:AT1G22930 T-complex protein 11
Length=1020
Score = 29.3 bits (64), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 16 DNALKALLLQLPDWSTLSTHMKTIDPEALHLAIRSVKADV 55
DN ++++ L+ PD+S +S MK + E + S K ++
Sbjct 586 DNVMESMKLEKPDYSCISNLMKEVSDELCQMVPDSWKVEI 625
> ath:AT2G35860 FLA16; FLA16 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN
16 PRECURSOR)
Length=445
Score = 29.3 bits (64), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 20/75 (26%), Positives = 35/75 (46%), Gaps = 13/75 (17%)
Query 27 PDWSTLSTHMKTIDPEALHLA--IRSVKADVAAALKTE-------MAKEYEHLTLPAGEE 77
P W +LS H +T+ + LHL + ++K D A ++ + + E L +P +
Sbjct 120 PQWPSLSHHHRTLSNDHLHLTVDVNTLKVDSAEIIRPDDVIRPDGIIHGIERLLIPRSVQ 179
Query 78 DDMEEESTGRRKLRN 92
+D RR LR+
Sbjct 180 EDFNR----RRSLRS 190
> hsa:89910 UBE3B, DKFZp586K2123, DKFZp686A1051, FLJ45294, MGC131858,
MGC78388; ubiquitin protein ligase E3B (EC:6.3.2.-);
K10588 ubiquitin-protein ligase E3 B [EC:6.3.2.19]
Length=1068
Score = 28.9 bits (63), Expect = 6.9, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 1 AYIDALRTTLTDNTLDNALKALLLQLPDWSTLSTHMKTIDPEAL 44
A+ ALR + DN ++ L+ + L TH+ T+ PE L
Sbjct 232 AFSLALRPVIAAQFSDNLIRPFLIHIMSVPALVTHLSTVTPERL 275
> hsa:319089 TTC6, C14orf25, MGC119358, MGC119360, MGC119361;
tetratricopeptide repeat domain 6
Length=1770
Score = 28.5 bits (62), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 33/62 (53%), Gaps = 3/62 (4%)
Query 49 RSVKADVAAALKTEMAKEYEHLTLPAGEEDDMEEESTGRRKLRNTLLFFLSESRDREAVD 108
R +VA MAKE + ++ A ED E S+GRRK+R F+SES REA +
Sbjct 341 REQSQEVAPLAGPCMAKERKASSVSA--EDGYMEASSGRRKVR-IRSNFVSESGAREARE 397
Query 109 RA 110
A
Sbjct 398 AA 399
> pfa:PFL2195w clathrin coat assembly protein AP180, putative
Length=431
Score = 28.5 bits (62), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 11/43 (25%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 90 LRNTLLFFLSESRDREAVDRAVKHFTDAKCMTDRYAGLITLGN 132
++N +F+S+ +D+E + + + HFT + + D+ G+ + N
Sbjct 92 MKNGCEYFISDVKDKEELIKKLTHFTHLEDLKDKGIGIRDISN 134
> bbo:BBOV_IV008540 23.m06569; helicase
Length=317
Score = 28.5 bits (62), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 23/94 (24%), Positives = 34/94 (36%), Gaps = 1/94 (1%)
Query 34 THMKTIDPEALHLAIRSVKADVAAALKTEMAKEYEHLTLPAGEEDDMEEESTGRRKLRNT 93
T +KT+DP+ + I A ++ + E P D + R + N
Sbjct 209 TQLKTLDPQTGNRYIVCTDA-ISRGIDIENISLVVQFDFPKNVLDYIHRAGRAARNMLNG 267
Query 94 LLFFLSESRDREAVDRAVKHFTDAKCMTDRYAGL 127
L E DRE + KH D + R GL
Sbjct 268 RTVLLWEDGDREFCELLNKHMNDLPSLFSRKRGL 301
Lambda K H
0.317 0.130 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3134054208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40