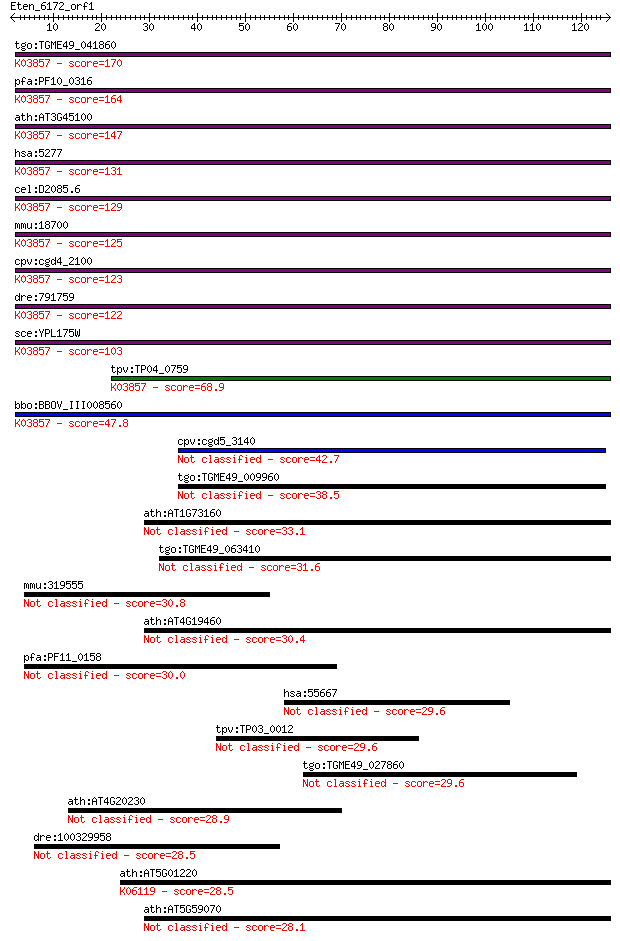
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6172_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_041860 phosphatidylinositolglycan class A protein (... 170 1e-42
pfa:PF10_0316 N-acetylglucosaminyl-phosphatidylinositol biosyn... 164 5e-41
ath:AT3G45100 SETH2; SETH2; transferase, transferring glycosyl... 147 8e-36
hsa:5277 PIGA, GPI3, PIG-A; phosphatidylinositol glycan anchor... 131 4e-31
cel:D2085.6 hypothetical protein; K03857 phosphatidylinositol ... 129 3e-30
mmu:18700 Piga, AI194334, Pig-a; phosphatidylinositol glycan a... 125 4e-29
cpv:cgd4_2100 PIG-A like N-acetylglucosaminyl-phosphatidylinos... 123 1e-28
dre:791759 piga, MGC56589, im:6911675, zgc:56589; phosphatidyl... 122 3e-28
sce:YPL175W SPT14, CWH6, GPI3; Spt14p (EC:2.4.1.198); K03857 p... 103 2e-22
tpv:TP04_0759 N-acetylglucosaminyl-phosphatidylinositol transf... 68.9 4e-12
bbo:BBOV_III008560 17.m07749; glycosyl transferase; K03857 pho... 47.8 8e-06
cpv:cgd5_3140 LPS glycosyltransferase of possible cyanobacteri... 42.7 3e-04
tgo:TGME49_009960 glycan synthetase, putative (EC:2.4.1.21 2.4... 38.5 0.005
ath:AT1G73160 glycosyl transferase family 1 protein 33.1 0.20
tgo:TGME49_063410 scavenger receptor protein SR2 (EC:3.4.21.84... 31.6 0.74
mmu:319555 Nwd1, A230063L24Rik; NACHT and WD repeat domain con... 30.8 1.1
ath:AT4G19460 glycosyl transferase family 1 protein 30.4 1.4
pfa:PF11_0158 conserved Plasmodium protein 30.0 2.1
hsa:55667 DENND4C, C9orf55, C9orf55B, DKFZp686I09113, FLJ20686... 29.6 2.3
tpv:TP03_0012 hypothetical protein 29.6 2.4
tgo:TGME49_027860 hypothetical protein 29.6 2.5
ath:AT4G20230 terpene synthase/cyclase family protein 28.9 3.9
dre:100329958 LReO_3-like 28.5 4.9
ath:AT5G01220 SQD2; SQD2 (sulfoquinovosyldiacylglycerol 2); UD... 28.5 6.1
ath:AT5G59070 glycosyl transferase family 1 protein 28.1 6.9
> tgo:TGME49_041860 phosphatidylinositolglycan class A protein
(EC:2.4.1.198); K03857 phosphatidylinositol glycan, class A
[EC:2.4.1.198]
Length=616
Score = 170 bits (430), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 79/124 (63%), Positives = 94/124 (75%), Gaps = 0/124 (0%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
LG VVYTDHS+F FAD ACIH+NK+ +F L D CICVSHTH+EN VLR + P ++
Sbjct 133 LGMHVVYTDHSLFGFADMACIHLNKVLRFVLHDLDACICVSHTHRENFVLRAGVPPSRVY 192
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
VINNAVD VPD SKRP PP I +++LSRL+YRKGIDLLV +IP +C K PNV F+IG
Sbjct 193 VINNAVDASTLVPDPSKRPKPPEIRVVVLSRLTYRKGIDLLVTVIPPICKKLPNVNFVIG 252
Query 122 GDGP 125
G GP
Sbjct 253 GYGP 256
> pfa:PF10_0316 N-acetylglucosaminyl-phosphatidylinositol biosynthetic
protein, putative; K03857 phosphatidylinositol glycan,
class A [EC:2.4.1.198]
Length=461
Score = 164 bits (416), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 72/124 (58%), Positives = 97/124 (78%), Gaps = 0/124 (0%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
LG + +YTDHS++SF+D CIH+NK+ K+ + DH ICVSHT++ENLVLR NP++ +
Sbjct 139 LGIKTIYTDHSLYSFSDKGCIHVNKLLKYCINDVDHSICVSHTNRENLVLRTESNPYKTS 198
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
VI NA+D +VP +SKRP P INI+++SRL+YRKGIDL+V +IP VC KYP + FIIG
Sbjct 199 VIGNALDTTKFVPCISKRPKFPRINIIVISRLTYRKGIDLIVKVIPLVCQKYPFIKFIIG 258
Query 122 GDGP 125
G+GP
Sbjct 259 GEGP 262
> ath:AT3G45100 SETH2; SETH2; transferase, transferring glycosyl
groups; K03857 phosphatidylinositol glycan, class A [EC:2.4.1.198]
Length=447
Score = 147 bits (371), Expect = 8e-36, Method: Composition-based stats.
Identities = 69/124 (55%), Positives = 88/124 (70%), Gaps = 1/124 (0%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+GY+VV+TDHS++ FAD IHMNK+ +F L D ICVSHT KEN VLR L+P ++
Sbjct 119 MGYKVVFTDHSLYGFADVGSIHMNKVLQFSLADIDQAICVSHTSKENTVLRSGLSPAKVF 178
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
+I NAVD + P S RP I I+++SRL YRKG DLLV++IP VC YPNV F++G
Sbjct 179 MIPNAVDTAMFKP-ASVRPSTDIITIVVISRLVYRKGADLLVEVIPEVCRLYPNVRFVVG 237
Query 122 GDGP 125
GDGP
Sbjct 238 GDGP 241
> hsa:5277 PIGA, GPI3, PIG-A; phosphatidylinositol glycan anchor
biosynthesis, class A (EC:2.4.1.198); K03857 phosphatidylinositol
glycan, class A [EC:2.4.1.198]
Length=484
Score = 131 bits (330), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 65/124 (52%), Positives = 86/124 (69%), Gaps = 2/124 (1%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+G + V+TDHS+F FAD + + NK+ L +H ICVS+T KEN VLR ALNP ++
Sbjct 145 MGLQTVFTDHSLFGFADVSSVLTNKLLTVSLCDTNHIICVSYTSKENTVLRAALNPEIVS 204
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
VI NAVD + PD +R +I I+++SRL YRKGIDLL IIP +C KYP++ FIIG
Sbjct 205 VIPNAVDPTDFTPDPFRR--HDSITIVVVSRLVYRKGIDLLSGIIPELCQKYPDLNFIIG 262
Query 122 GDGP 125
G+GP
Sbjct 263 GEGP 266
> cel:D2085.6 hypothetical protein; K03857 phosphatidylinositol
glycan, class A [EC:2.4.1.198]
Length=444
Score = 129 bits (323), Expect = 3e-30, Method: Composition-based stats.
Identities = 62/125 (49%), Positives = 87/125 (69%), Gaps = 2/125 (1%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNK-IFKFFLTHCDHCICVSHTHKENLVLRGALNPHQI 60
+G R V+TDHS+F FADA+ I NK + ++ L + D ICVS+T KEN VLRG L+P+++
Sbjct 120 MGLRTVFTDHSLFGFADASAILTNKLVLQYSLINVDQTICVSYTSKENTVLRGKLDPNKV 179
Query 61 AVINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFII 120
+ I NA++ + PD ++ P I+ L RL YRKG DLL +I+P VCA++ +V FII
Sbjct 180 STIPNAIETSLFTPDRNQFFNNPT-TIVFLGRLVYRKGADLLCEIVPKVCARHKSVRFII 238
Query 121 GGDGP 125
GGDGP
Sbjct 239 GGDGP 243
> mmu:18700 Piga, AI194334, Pig-a; phosphatidylinositol glycan
anchor biosynthesis, class A (EC:2.4.1.198); K03857 phosphatidylinositol
glycan, class A [EC:2.4.1.198]
Length=485
Score = 125 bits (313), Expect = 4e-29, Method: Composition-based stats.
Identities = 61/124 (49%), Positives = 82/124 (66%), Gaps = 1/124 (0%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+G + V+TDHS+F FAD + + NK+ L +H ICVS+T KEN VLR ALNP ++
Sbjct 145 MGLQTVFTDHSLFGFADVSSVLTNKLLTVSLCDTNHIICVSYTSKENTVLRAALNPEIVS 204
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
VI NAVD + PD +R I ++++SRL YRKG DLL IIP +C KY + F+IG
Sbjct 205 VIPNAVDPTDFTPDPFRR-HDSVITVVVVSRLVYRKGTDLLSGIIPELCQKYQELHFLIG 263
Query 122 GDGP 125
G+GP
Sbjct 264 GEGP 267
> cpv:cgd4_2100 PIG-A like N-acetylglucosaminyl-phosphatidylinositol
biosynthetic protein ; K03857 phosphatidylinositol glycan,
class A [EC:2.4.1.198]
Length=451
Score = 123 bits (309), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 58/125 (46%), Positives = 87/125 (69%), Gaps = 1/125 (0%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+GYRVV+TDHS+F ++ IH N F+ L+ D+ ICVSHT+K+NL+ R ++P +
Sbjct 134 MGYRVVFTDHSLFGLSNYDSIHANNFFRINLSCIDNVICVSHTNKKNLIYRSLISPKNVT 193
Query 62 VINNAVDVPAYVPDLS-KRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFII 120
VI NA+D ++P+ + K P + ++ + R++YRKG+DLLV+IIP +C PNV FI+
Sbjct 194 VIPNAIDSNDFLPNPNYKSPINNEVIVISICRMTYRKGVDLLVEIIPRICNSDPNVRFIL 253
Query 121 GGDGP 125
GGDGP
Sbjct 254 GGDGP 258
> dre:791759 piga, MGC56589, im:6911675, zgc:56589; phosphatidylinositol
glycan anchor biosynthesis, class A (EC:2.4.1.198);
K03857 phosphatidylinositol glycan, class A [EC:2.4.1.198]
Length=487
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 59/124 (47%), Positives = 83/124 (66%), Gaps = 1/124 (0%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+G V+TDHS+F FAD + + NK+ L +H +CVS+T KEN VLR L+P ++
Sbjct 149 MGLNTVFTDHSLFGFADLSSVLTNKLLTVSLCDTNHIVCVSYTSKENTVLRATLDPEIVS 208
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
VI NAVD + PD +R I I+++SRL YRKGIDLL IIP +C ++P++ F+IG
Sbjct 209 VIPNAVDPTDFTPDPCRR-DNSKITIVVISRLVYRKGIDLLSGIIPELCGRHPDLCFLIG 267
Query 122 GDGP 125
G+GP
Sbjct 268 GEGP 271
> sce:YPL175W SPT14, CWH6, GPI3; Spt14p (EC:2.4.1.198); K03857
phosphatidylinositol glycan, class A [EC:2.4.1.198]
Length=452
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 54/129 (41%), Positives = 78/129 (60%), Gaps = 5/129 (3%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+G R V+TDHS++ F + I +NK+ F LT+ D ICVS+T KEN+++R L+P I+
Sbjct 115 MGLRTVFTDHSLYGFNNLTSIWVNKLLTFTLTNIDRVICVSNTCKENMIVRTELSPDIIS 174
Query 62 VINNAVDVPAYVP-----DLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNV 116
VI NAV + P ++ I I+++ RL KG DLL IIP VC+ + +V
Sbjct 175 VIPNAVVSEDFKPRDPTGGTKRKQSRDKIVIVVIGRLFPNKGSDLLTRIIPKVCSSHEDV 234
Query 117 TFIIGGDGP 125
FI+ GDGP
Sbjct 235 EFIVAGDGP 243
> tpv:TP04_0759 N-acetylglucosaminyl-phosphatidylinositol transferase;
K03857 phosphatidylinositol glycan, class A [EC:2.4.1.198]
Length=400
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 45/130 (34%), Positives = 65/130 (50%), Gaps = 26/130 (20%)
Query 22 IHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIAVINNA------------VDV 69
IH++ + F D+ I VS+ H+EN+VLR +P +++VI NA +++
Sbjct 102 IHIHMYMRVFTMFEDNIIAVSNKHRENMVLRSYADPRKVSVIPNALVCFCLFYFYYLINL 161
Query 70 P-----------AYVPDLSKRPPPPA---INILILSRLSYRKGIDLLVDIIPAVCAKYPN 115
P D P + I I+ +SRL RKGIDLL ++P VC + N
Sbjct 162 PYLHIHLNTIFMQESDDFKCNDEPLSKDKIVIVYISRLCERKGIDLLTQVVPLVCKNHDN 221
Query 116 VTFIIGGDGP 125
V FIIGG GP
Sbjct 222 VEFIIGGGGP 231
> bbo:BBOV_III008560 17.m07749; glycosyl transferase; K03857 phosphatidylinositol
glycan, class A [EC:2.4.1.198]
Length=397
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 38/124 (30%), Positives = 54/124 (43%), Gaps = 42/124 (33%)
Query 2 LGYRVVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIA 61
+G + V+TDHS+FSF D L P +
Sbjct 122 MGLKTVFTDHSLFSFID------------------------------------LGPTILT 145
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIG 121
+ ++A P P + I I+++SRL+ KG LL +IP VC ++ NV FIIG
Sbjct 146 LESDAFK-PRSEPRNDDK-----IVIVVISRLTAIKGAMLLNAVIPIVCHRHANVNFIIG 199
Query 122 GDGP 125
GDGP
Sbjct 200 GDGP 203
> cpv:cgd5_3140 LPS glycosyltransferase of possible cyanobacterial
origin
Length=2069
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 9/98 (9%)
Query 36 DHCICVSHTHKENLVLRGALNPHQIAVINNAVDVPAY----VPDLS--KRP---PPPAIN 86
D ICVS E + +N ++I VI N ++ + + D KR P
Sbjct 1402 DRLICVSGVFAEEVCRLFGINRNKIKVIFNGINCKTFDSVAMDDAGEVKRQYGIGPLEPT 1461
Query 87 ILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIGGDG 124
L ++R++ +KG+DLL++ +P + + FII GDG
Sbjct 1462 FLCVARMALQKGVDLLIEAVPGILKYRNDTKFIIVGDG 1499
> tgo:TGME49_009960 glycan synthetase, putative (EC:2.4.1.21 2.4.1.18)
Length=1707
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 25/96 (26%), Positives = 46/96 (47%), Gaps = 7/96 (7%)
Query 36 DHCICVSHTHKENLVLRGALNPHQIAVINNAVDVPAY-----VPDLSKRPPPPAIN--IL 88
D ICVS + + + ++P +I VI N + + ++ + PA++ L
Sbjct 1210 DRVICVSGVLAQEVQTQYGIHPEKIKVIYNGIQCERFDGEVDAGEVKAQYGIPAMDPTFL 1269
Query 89 ILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIGGDG 124
+ R+ +KG DLL++ IP + + F+ GDG
Sbjct 1270 FVGRMVVQKGPDLLLEAIPFILKFRSDAKFVFVGDG 1305
> ath:AT1G73160 glycosyl transferase family 1 protein
Length=486
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 31/107 (28%), Positives = 48/107 (44%), Gaps = 11/107 (10%)
Query 29 KFFLTHCDHCICVSHTHKENLVLRGALNPHQIAVINNAVDVPAYV--PDLSKR------P 80
+FF + H IC+S++ +E LV L ++ VI N VD +V P+ R
Sbjct 235 RFFPKYKQH-ICISNSAREVLVNIYQLPKRKVHVIVNGVDQTKFVYSPESGARFRAKHGI 293
Query 81 PPPAINIL--ILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIGGDGP 125
P I+ + RL KG LL + + +P V ++ G GP
Sbjct 294 PDNGTYIVMGVSGRLVRDKGHPLLYEAFALLVKMHPKVYLLVAGSGP 340
> tgo:TGME49_063410 scavenger receptor protein SR2 (EC:3.4.21.84
1.13.11.40)
Length=1250
Score = 31.6 bits (70), Expect = 0.74, Method: Composition-based stats.
Identities = 30/106 (28%), Positives = 47/106 (44%), Gaps = 17/106 (16%)
Query 32 LTHCDHCIC--VSHTHKENLVL--RGALNPHQIAVINNAVDVPAYVPDLSKRPPPPAINI 87
L HC H I + H+E++++ RG +P + + D+PA LSKR PP I +
Sbjct 609 LRHCPHEISSDIYCVHEEDIIVGCRGDGDPSGMGLFRTE-DIPA----LSKRSLPPKIEL 663
Query 88 LILSRLSYRKGID-----LLVDIIPAVCAKYPNV---TFIIGGDGP 125
R +K + + V P C++ P T+I D P
Sbjct 664 NCGDRPLSQKQMGGQAGAMFVATCPEGCSEEPGSLKGTYIYTDDSP 709
> mmu:319555 Nwd1, A230063L24Rik; NACHT and WD repeat domain containing
1
Length=1521
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 28/53 (52%), Gaps = 7/53 (13%)
Query 4 YRVVY--TDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGA 54
YRVVY +D S+F + D AC +K+F H CV +H E L + GA
Sbjct 1318 YRVVYGMSDGSLFLY-DCAC---SKVFPL-EAHGSRVSCVEVSHSEQLAVSGA 1365
> ath:AT4G19460 glycosyl transferase family 1 protein
Length=516
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 29/107 (27%), Positives = 44/107 (41%), Gaps = 11/107 (10%)
Query 29 KFFLTHCDHCICVSHTHKENLVLRGALNPHQIAVINNAVDVPAYVPD-------LSKRPP 81
+FF + H I +S + E L + ++ VI N VD + D SK
Sbjct 270 RFFHNYAHH-IAISDSCGEMLRDVYQIPEKRVHVILNGVDENGFTSDKKLRTLFRSKLGL 328
Query 82 PPAINILILS---RLSYRKGIDLLVDIIPAVCAKYPNVTFIIGGDGP 125
P + ++L RL KG LL + + Y NV ++ G GP
Sbjct 329 PENSSAIVLGAAGRLVKDKGHPLLFEAFAKIIQTYSNVYLVVAGSGP 375
> pfa:PF11_0158 conserved Plasmodium protein
Length=2480
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 18/68 (26%), Positives = 29/68 (42%), Gaps = 11/68 (16%)
Query 4 YRVVYTDHSMFSFADAACIHMNKIFKFF---LTHCDHCICVSHTHKENLVLRGALNPHQI 60
Y +Y D ++ F ++MNK K++ CD+ I H KE N H +
Sbjct 291 YLYIYDDGIIYVFVSIVGVNMNKFQKYYYDNFVQCDNNIYNDHDKKE--------NEHSL 342
Query 61 AVINNAVD 68
+ NN +
Sbjct 343 YLTNNKTN 350
> hsa:55667 DENND4C, C9orf55, C9orf55B, DKFZp686I09113, FLJ20686,
bA513M16.3; DENN/MADD domain containing 4C
Length=1673
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 58 HQIAVINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVD 104
H I+ ++ + V L KRP PP +++ LS L RK ++ L++
Sbjct 1420 HGISTVSLPNSLQEVVDPLGKRPNPPPVSVPYLSPLVLRKELESLLE 1466
> tpv:TP03_0012 hypothetical protein
Length=512
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 44 THKENLVLRGALNPHQIAVINNAVDVPAYVPDLSKRPPPPAI 85
T+++ GAL Q +N +P YV DLS+R P AI
Sbjct 218 TYEKQFEETGALKAAQNCTYSNLSGIPRYVWDLSQRDPRHAI 259
> tgo:TGME49_027860 hypothetical protein
Length=377
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 26/57 (45%), Gaps = 4/57 (7%)
Query 62 VINNAVDVPAYVPDLSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTF 118
V VD P ++ + PP N+ ++ R + K + L AV KYPN+ F
Sbjct 29 VAQGLVDAPLWLSAAERAPPMELSNLKLMDRKVHNKYLSL----TQAVLKKYPNMRF 81
> ath:AT4G20230 terpene synthase/cyclase family protein
Length=609
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 5/57 (8%)
Query 13 MFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIAVINNAVDV 69
M S +AA H+ + L D + + +H E LV G PH +A I NA+ +
Sbjct 202 MLSLYEAA--HLATTKDYIL---DEALIFTSSHLETLVATGTCPPHLLARIRNALSI 253
> dre:100329958 LReO_3-like
Length=843
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 6 VVYTDHSMFSFADAACIHMNKIFKFFLTHCDHCICVSHTHKENLVLRGALN 56
VVYTDH+ F + C H ++ ++ L + + + H VL AL+
Sbjct 791 VVYTDHNPLVFLNRMCNHNQRLMRWALVTQGYNLEIRHKKGSENVLADALS 841
> ath:AT5G01220 SQD2; SQD2 (sulfoquinovosyldiacylglycerol 2);
UDP-glycosyltransferase/ UDP-sulfoquinovose:DAG sulfoquinovosyltransferase/
transferase, transferring glycosyl groups; K06119
sulfoquinovosyltransferase [EC:2.4.1.-]
Length=510
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 27/111 (24%), Positives = 45/111 (40%), Gaps = 14/111 (12%)
Query 24 MNKIFKFFLTHCDHCICVSHTHKENLVLRGALNPHQIAVINNAVDVPAYVPD-------- 75
M I +F D + S ++L+ GA +Q+ + N VD ++ P
Sbjct 240 MWSIIRFLHRAADLTLVPSAAIGKDLIAAGATAANQLRLWNKGVDSESFNPRFRSQEMRI 299
Query 76 -LSKRPPPPAINILILSRLSYRKGIDLLVDIIPAVCAKYPNVTFIIGGDGP 125
LS P + ++ + R+ K ++LL +V K P GDGP
Sbjct 300 RLSNGEPEKPL-VIHVGRIGVEKSLELL----KSVMDKLPEARIAFIGDGP 345
> ath:AT5G59070 glycosyl transferase family 1 protein
Length=505
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 29/115 (25%), Positives = 47/115 (40%), Gaps = 20/115 (17%)
Query 29 KFFLTHCDHCICVSHTHKENLVLRGALNPHQ-IAVINNAVDVPAYVPDLSKRPP------ 81
KFF + H + H +++ R + P + + +I N VD + PD+SKR
Sbjct 250 KFFQRYAHHV--ATSDHCGDVLKRIYMIPEERVHIILNGVDENVFKPDVSKRESFREKFG 307
Query 82 -------PPAINILILSRLSYRKGIDLLVDIIPAVCAK----YPNVTFIIGGDGP 125
+ + I RL KG L+ + V + NV ++ GDGP
Sbjct 308 VRSGKNKKSPLVLGIAGRLVRDKGHPLMFSALKRVFEENKEARENVVVLVAGDGP 362
Lambda K H
0.328 0.144 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064871684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40