bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6168_orf1
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
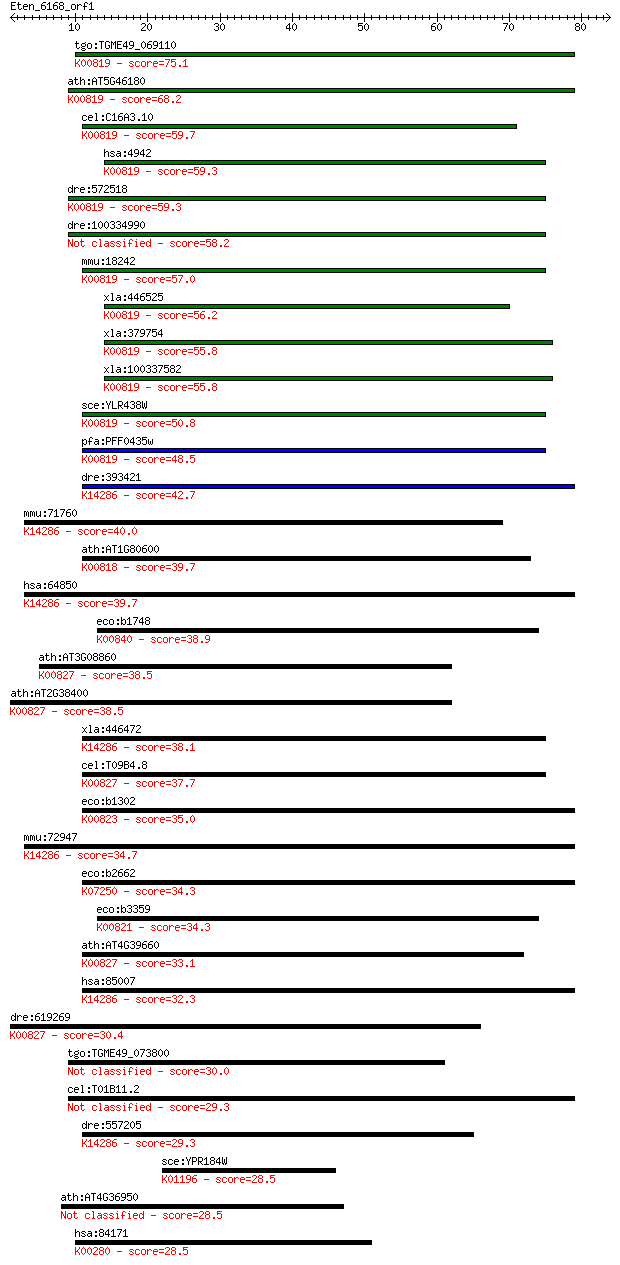
tgo:TGME49_069110 ornithine aminotransferase, putative (EC:2.6... 75.1 6e-14
ath:AT5G46180 delta-OAT; ornithine-oxo-acid transaminase (EC:2... 68.2 7e-12
cel:C16A3.10 hypothetical protein; K00819 ornithine--oxo-acid ... 59.7 2e-09
hsa:4942 OAT, DKFZp781A11155, GACR, HOGA, OATASE, OKT; ornithi... 59.3 3e-09
dre:572518 Ornithine aminotransferase, mitochondrial-like; K00... 59.3 3e-09
dre:100334990 Ornithine aminotransferase, mitochondrial-like 58.2 6e-09
mmu:18242 Oat, AI194874; ornithine aminotransferase (EC:2.6.1.... 57.0 2e-08
xla:446525 oat, MGC80244; ornithine aminotransferase (gyrate a... 56.2 3e-08
xla:379754 oat, MGC53882, hoga; ornithine aminotransferase (EC... 55.8 3e-08
xla:100337582 oat; ornithine aminotransferase (EC:2.6.1.13); K... 55.8 3e-08
sce:YLR438W CAR2; Car2p (EC:2.6.1.13); K00819 ornithine--oxo-a... 50.8 1e-06
pfa:PFF0435w ornithine aminotransferase (EC:2.6.1.13); K00819 ... 48.5 5e-06
dre:393421 agxt2l1, MGC63486, zgc:63486; alanine-glyoxylate am... 42.7 3e-04
mmu:71760 Agxt2l1, 1300019H02Rik, AI195447; alanine-glyoxylate... 40.0 0.002
ath:AT1G80600 WIN1; WIN1 (HOPW1-1-INTERACTING 1); N2-acetyl-L-... 39.7 0.002
hsa:64850 AGXT2L1; alanine-glyoxylate aminotransferase 2-like ... 39.7 0.003
eco:b1748 astC, argM, cstC, ECK1746, JW1737, ydjW; succinylorn... 38.9 0.004
ath:AT3G08860 alanine--glyoxylate aminotransferase, putative /... 38.5 0.005
ath:AT2G38400 AGT3; AGT3 (ALANINE:GLYOXYLATE AMINOTRANSFERASE ... 38.5 0.005
xla:446472 agxt2l1, MGC79033; alanine-glyoxylate aminotransfer... 38.1 0.007
cel:T09B4.8 hypothetical protein; K00827 alanine-glyoxylate tr... 37.7 0.009
eco:b1302 puuE, ECK1297, goaG, JW1295; GABA aminotransferase, ... 35.0 0.058
mmu:72947 Agxt2l2, 2900006B13Rik; alanine-glyoxylate aminotran... 34.7 0.073
eco:b2662 gabT, ECK2656, JW2637; 4-aminobutyrate aminotransfer... 34.3 0.096
eco:b3359 argD, alaB, arg1, argG, dapC, dtu, ECK3347, JW3322; ... 34.3 0.100
ath:AT4G39660 AGT2; AGT2 (ALANINE:GLYOXYLATE AMINOTRANSFERASE ... 33.1 0.22
hsa:85007 AGXT2L2, MGC117348, MGC15875, MGC45484; alanine-glyo... 32.3 0.37
dre:619269 agxt2, MGC114195, im:7153274, zgc:114195; alanine-g... 30.4 1.6
tgo:TGME49_073800 hypothetical protein 30.0 2.1
cel:T01B11.2 hypothetical protein 29.3 3.4
dre:557205 MGC123007; zgc:123007; K14286 alanine-glyoxylate am... 29.3 3.5
sce:YPR184W GDB1; Gdb1p (EC:3.2.1.33 2.4.1.25); K01196 glycoge... 28.5 5.2
ath:AT4G36950 MAPKKK21; ATP binding / protein kinase/ protein ... 28.5 5.4
hsa:84171 LOXL4, FLJ21889, LOXC; lysyl oxidase-like 4; K00280 ... 28.5 5.9
> tgo:TGME49_069110 ornithine aminotransferase, putative (EC:2.6.1.13);
K00819 ornithine--oxo-acid transaminase [EC:2.6.1.13]
Length=441
Score = 75.1 bits (183), Expect = 6e-14, Method: Composition-based stats.
Identities = 32/69 (46%), Positives = 48/69 (69%), Gaps = 0/69 (0%)
Query 10 WIRDIRGRGLFNAVEINGSSGEAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKD 69
WI++IRGRGL NAVE++ + + + ++LK G+L +PTRG V+RF+PPL I + + +D
Sbjct 364 WIKEIRGRGLLNAVEVDSDAIDPNDVVMKLKENGILSKPTRGRVMRFIPPLVITDEEHRD 423
Query 70 ALQRIQDVF 78
A RI F
Sbjct 424 ATTRIIKSF 432
> ath:AT5G46180 delta-OAT; ornithine-oxo-acid transaminase (EC:2.6.1.13);
K00819 ornithine--oxo-acid transaminase [EC:2.6.1.13]
Length=475
Score = 68.2 bits (165), Expect = 7e-12, Method: Composition-based stats.
Identities = 29/73 (39%), Positives = 45/73 (61%), Gaps = 3/73 (4%)
Query 9 EWIRDIRGRGLFNAVEINGSS---GEAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEA 65
++I+++RGRGLFNA+E N S A +C+ LK GVL +PT ++R PPL I
Sbjct 371 KYIKEVRGRGLFNAIEFNSESLSPVSAYDICLSLKERGVLAKPTHNTIVRLTPPLSISSD 430
Query 66 QMKDALQRIQDVF 78
+++D + + DV
Sbjct 431 ELRDGSEALHDVL 443
> cel:C16A3.10 hypothetical protein; K00819 ornithine--oxo-acid
transaminase [EC:2.6.1.13]
Length=357
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 25/60 (41%), Positives = 43/60 (71%), Gaps = 1/60 (1%)
Query 11 IRDIRGRGLFNAVEINGSSGEAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDA 70
+ +RG+GLF A+ IN +A ++C++LK G+L + T G+++RF PPLCI++ Q++ A
Sbjct 284 VSTVRGKGLFCAIVINKKY-DAWKVCLKLKENGLLAKNTHGDIIRFAPPLCINKEQVEQA 342
> hsa:4942 OAT, DKFZp781A11155, GACR, HOGA, OATASE, OKT; ornithine
aminotransferase (EC:2.6.1.13); K00819 ornithine--oxo-acid
transaminase [EC:2.6.1.13]
Length=439
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 45/62 (72%), Gaps = 1/62 (1%)
Query 14 IRGRGLFNAVEINGSSG-EAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDALQ 72
+RG+GL NA+ I + +A ++C+ L++ G+L +PT G+++RF PPL I E +++++++
Sbjct 371 VRGKGLLNAIVIKETKDWDAWKVCLRLRDNGLLAKPTHGDIIRFAPPLVIKEDELRESIE 430
Query 73 RI 74
I
Sbjct 431 II 432
> dre:572518 Ornithine aminotransferase, mitochondrial-like; K00819
ornithine--oxo-acid transaminase [EC:2.6.1.13]
Length=444
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 46/67 (68%), Gaps = 1/67 (1%)
Query 9 EWIRDIRGRGLFNAVEINGSSG-EAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQM 67
E + +RG+GL NA+ I + +A Q+C+ L++ G+L +PT G+++R PPL I+E ++
Sbjct 371 EIVSGVRGKGLLNAIIIKETKDYDAWQVCLRLRDNGLLAKPTHGDIIRLAPPLTINEQEV 430
Query 68 KDALQRI 74
++ ++ I
Sbjct 431 RECVEII 437
> dre:100334990 Ornithine aminotransferase, mitochondrial-like
Length=265
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 46/67 (68%), Gaps = 1/67 (1%)
Query 9 EWIRDIRGRGLFNAVEINGSSG-EAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQM 67
E + +RG+GL NA+ I + +A Q+C+ L++ G+L +PT G+++R PPL I+E ++
Sbjct 192 EIVSGVRGKGLLNAIIIKETKDYDAWQVCLRLRDNGLLAKPTHGDIIRLAPPLTINEQEV 251
Query 68 KDALQRI 74
++ ++ I
Sbjct 252 RECVEII 258
> mmu:18242 Oat, AI194874; ornithine aminotransferase (EC:2.6.1.13);
K00819 ornithine--oxo-acid transaminase [EC:2.6.1.13]
Length=439
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 45/65 (69%), Gaps = 1/65 (1%)
Query 11 IRDIRGRGLFNAVEINGSSG-EAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKD 69
+ +RG+GL NA+ I + +A ++C+ L++ G+L +PT G+++R PPL I E ++++
Sbjct 368 VTSVRGKGLLNAIVIRETKDCDAWKVCLRLRDNGLLAKPTHGDIIRLAPPLVIKEDEIRE 427
Query 70 ALQRI 74
+++ I
Sbjct 428 SVEII 432
> xla:446525 oat, MGC80244; ornithine aminotransferase (gyrate
atrophy) (EC:2.6.1.13); K00819 ornithine--oxo-acid transaminase
[EC:2.6.1.13]
Length=439
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/57 (40%), Positives = 40/57 (70%), Gaps = 1/57 (1%)
Query 14 IRGRGLFNAVEINGSSG-EAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKD 69
+RG+GL NAV I S +A ++C+ L++ G+L +PT G+++R PPL I E ++++
Sbjct 371 VRGKGLLNAVVIKQSKDCDAWKVCLRLRDNGLLAKPTHGDIIRLAPPLTIKEDEIRE 427
> xla:379754 oat, MGC53882, hoga; ornithine aminotransferase (EC:2.6.1.13);
K00819 ornithine--oxo-acid transaminase [EC:2.6.1.13]
Length=439
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 42/63 (66%), Gaps = 1/63 (1%)
Query 14 IRGRGLFNAVEINGSSG-EAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDALQ 72
+RG+GL NA+ I S +A ++C+ L++ G+L +PT G+++R PPL I E ++++ +
Sbjct 371 VRGKGLLNAIVIKQSKDCDAWKVCLRLRDNGLLAKPTHGDIIRLAPPLTIKEDEIRECSE 430
Query 73 RIQ 75
I
Sbjct 431 IIH 433
> xla:100337582 oat; ornithine aminotransferase (EC:2.6.1.13);
K00819 ornithine--oxo-acid transaminase [EC:2.6.1.13]
Length=439
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/63 (36%), Positives = 42/63 (66%), Gaps = 1/63 (1%)
Query 14 IRGRGLFNAVEINGSSG-EAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDALQ 72
+RG+GL NA+ I S +A ++C+ L++ G+L +PT G+++R PPL I E ++++ +
Sbjct 371 VRGKGLLNAIVIKQSKDCDAWKVCLRLRDNGLLAKPTHGDIIRLAPPLTIKEDEIRECSE 430
Query 73 RIQ 75
I
Sbjct 431 IIH 433
> sce:YLR438W CAR2; Car2p (EC:2.6.1.13); K00819 ornithine--oxo-acid
transaminase [EC:2.6.1.13]
Length=424
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 39/67 (58%), Gaps = 3/67 (4%)
Query 11 IRDIRGRGLFNAVEINGSSGE---AMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQM 67
I ++RG GL A+ I+ S A LC+ +K+ G+L +PT +++R PPL I E +
Sbjct 351 ISEVRGMGLLTAIVIDPSKANGKTAWDLCLLMKDHGLLAKPTHDHIIRLAPPLVISEEDL 410
Query 68 KDALQRI 74
+ ++ I
Sbjct 411 QTGVETI 417
> pfa:PFF0435w ornithine aminotransferase (EC:2.6.1.13); K00819
ornithine--oxo-acid transaminase [EC:2.6.1.13]
Length=414
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 34/64 (53%), Gaps = 0/64 (0%)
Query 11 IRDIRGRGLFNAVEINGSSGEAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDA 70
+R++RG+GL A+E +C++ K G++ R +R PPLCI + Q+ +
Sbjct 340 VREVRGKGLLCAIEFKNDLVNVWDICLKFKENGLITRSVHDKTVRLTPPLCITKEQLDEC 399
Query 71 LQRI 74
+ I
Sbjct 400 TEII 403
> dre:393421 agxt2l1, MGC63486, zgc:63486; alanine-glyoxylate
aminotransferase 2-like 1 (EC:2.6.1.-); K14286 alanine-glyoxylate
aminotransferase 2-like [EC:2.6.1.-]
Length=492
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 41/78 (52%), Gaps = 11/78 (14%)
Query 11 IRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCR---PTRGNVLRFLPPL 60
+ D+RGRGLF +E+ ++ EA ++ LK + +L P R NVL+F PP+
Sbjct 359 VGDVRGRGLFVGLELVRNQSKRTPATAEAQEVIYRLKEQRILLSADGPHR-NVLKFKPPM 417
Query 61 CIDEAQMKDALQRIQDVF 78
C + A+++I +
Sbjct 418 CFSREDAEFAVEKIDQIL 435
> mmu:71760 Agxt2l1, 1300019H02Rik, AI195447; alanine-glyoxylate
aminotransferase 2-like 1; K14286 alanine-glyoxylate aminotransferase
2-like [EC:2.6.1.-]
Length=499
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 26/76 (34%), Positives = 38/76 (50%), Gaps = 11/76 (14%)
Query 3 KEKCNLEWIRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCR---PTRGN 52
++K I DIRG GLF +++ ++ EA + E+K +GVL P R N
Sbjct 349 EQKAKHPLIGDIRGVGLFIGIDLVKDREKRTPATAEAQHIIYEMKGKGVLLSADGPHR-N 407
Query 53 VLRFLPPLCIDEAQMK 68
VL+ PP+C E K
Sbjct 408 VLKIKPPMCFTEDDAK 423
> ath:AT1G80600 WIN1; WIN1 (HOPW1-1-INTERACTING 1); N2-acetyl-L-ornithine:2-oxoglutarate
5-aminotransferase/ catalytic/ pyridoxal
phosphate binding / transaminase (EC:2.6.1.11); K00818
acetylornithine aminotransferase [EC:2.6.1.11]
Length=457
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 38/63 (60%), Gaps = 4/63 (6%)
Query 11 IRDIRGRGLFNAVEINGSSGEAMQLCIELKNEGVLCRPT-RGNVLRFLPPLCIDEAQMKD 69
++++RG GL VE++ + + C ++ G+L +GNV+R +PPL I E +++
Sbjct 388 VKEVRGEGLIIGVELDVPASSLVDAC---RDSGLLILTAGKGNVVRIVPPLVISEEEIER 444
Query 70 ALQ 72
A++
Sbjct 445 AVE 447
> hsa:64850 AGXT2L1; alanine-glyoxylate aminotransferase 2-like
1; K14286 alanine-glyoxylate aminotransferase 2-like [EC:2.6.1.-]
Length=493
Score = 39.7 bits (91), Expect = 0.003, Method: Composition-based stats.
Identities = 30/89 (33%), Positives = 42/89 (47%), Gaps = 14/89 (15%)
Query 3 KEKCNLEWIRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCR---PTRGN 52
K+K I DIRG GLF +++ ++ EA + ++K + VL P R N
Sbjct 343 KQKAKHTLIGDIRGIGLFIGIDLVKDHLKRTPATAEAQHIIYKMKEKRVLLSADGPHR-N 401
Query 53 VLRFLPPLCIDEAQMK---DALQRIQDVF 78
VL+ PP+C E K D L RI V
Sbjct 402 VLKIKPPMCFTEEDAKFMVDQLDRILTVL 430
> eco:b1748 astC, argM, cstC, ECK1746, JW1737, ydjW; succinylornithine
transaminase, PLP-dependent (EC:2.6.1.17 2.6.1.11);
K00840 succinylornithine aminotransferase [EC:2.6.1.81]
Length=406
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 32/62 (51%), Gaps = 1/62 (1%)
Query 13 DIRGRGLFNAVEINGS-SGEAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDAL 71
++RG GL +N +G+A Q+ E GV+ GNV+RF P L + E ++ L
Sbjct 331 EVRGLGLLIGCVLNADYAGQAKQISQEAAKAGVMVLIAGGNVVRFAPALNVSEEEVTTGL 390
Query 72 QR 73
R
Sbjct 391 DR 392
> ath:AT3G08860 alanine--glyoxylate aminotransferase, putative
/ beta-alanine-pyruvate aminotransferase, putative / AGT, putative;
K00827 alanine-glyoxylate transaminase / (R)-3-amino-2-methylpropionate-pyruvate
transaminase [EC:2.6.1.44 2.6.1.40]
Length=481
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 30/66 (45%), Gaps = 9/66 (13%)
Query 5 KCNLEWIRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCRPT--RGNVLR 55
K E I D+RGRGL VE + E + L ++K GVL GNV R
Sbjct 396 KNKYELIGDVRGRGLMLGVEFVKDRDLKTPAKAETLHLMDQMKEMGVLVGKGGFYGNVFR 455
Query 56 FLPPLC 61
PPLC
Sbjct 456 ITPPLC 461
> ath:AT2G38400 AGT3; AGT3 (ALANINE:GLYOXYLATE AMINOTRANSFERASE
3); alanine-glyoxylate transaminase/ catalytic/ pyridoxal
phosphate binding / transaminase; K00827 alanine-glyoxylate
transaminase / (R)-3-amino-2-methylpropionate-pyruvate transaminase
[EC:2.6.1.44 2.6.1.40]
Length=477
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 36/70 (51%), Gaps = 11/70 (15%)
Query 1 KMKEKCNLEWIRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCRPTR--G 51
++KEK E I D+RGRGL VE+ ++ E + + ++K GVL G
Sbjct 390 QLKEK--HEIIGDVRGRGLMLGVELVSDRKLKTPATAETLHIMDQMKELGVLIGKGGYFG 447
Query 52 NVLRFLPPLC 61
NV R PPLC
Sbjct 448 NVFRITPPLC 457
> xla:446472 agxt2l1, MGC79033; alanine-glyoxylate aminotransferase
2-like 1; K14286 alanine-glyoxylate aminotransferase 2-like
[EC:2.6.1.-]
Length=509
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 25/74 (33%), Positives = 38/74 (51%), Gaps = 11/74 (14%)
Query 11 IRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCR---PTRGNVLRFLPPL 60
I DIRG GLF V++ ++ EA + +LK + +L P R NVL+F PP+
Sbjct 358 IGDIRGVGLFVGVDLVKDRLFRTPATAEAQHIIYKLKEKRILLSADGPYR-NVLKFKPPM 416
Query 61 CIDEAQMKDALQRI 74
C ++ K + I
Sbjct 417 CFNKEDAKLVVDEI 430
> cel:T09B4.8 hypothetical protein; K00827 alanine-glyoxylate
transaminase / (R)-3-amino-2-methylpropionate-pyruvate transaminase
[EC:2.6.1.44 2.6.1.40]
Length=444
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 8/72 (11%)
Query 11 IRDIRGRGLFNAVEINGSSGEAMQ------LCIELKNEGVLCRP--TRGNVLRFLPPLCI 62
I D+RG+GL VE+ G+ + + + KN+G+L GNVLR PP+CI
Sbjct 365 IGDVRGKGLMIGVELIDEQGKPLAAAKTAAIFEDTKNQGLLIGKGGIHGNVLRIKPPMCI 424
Query 63 DEAQMKDALQRI 74
+ + A+ I
Sbjct 425 TKKDVDFAVDII 436
> eco:b1302 puuE, ECK1297, goaG, JW1295; GABA aminotransferase,
PLP-dependent (EC:2.6.1.19); K00823 4-aminobutyrate aminotransferase
[EC:2.6.1.19]
Length=421
Score = 35.0 bits (79), Expect = 0.058, Method: Composition-based stats.
Identities = 27/76 (35%), Positives = 38/76 (50%), Gaps = 8/76 (10%)
Query 11 IRDIRGRGLFNAVEIN----GSSGEAMQLCIELKN--EGVLCRP--TRGNVLRFLPPLCI 62
I +RG G AVE N G A+ I+ + +G+L GNV+RFL PL I
Sbjct 344 IAAVRGLGSMIAVEFNDPQTGEPSAAIAQKIQQRALAQGLLLLTCGAYGNVIRFLYPLTI 403
Query 63 DEAQMKDALQRIQDVF 78
+AQ A++ +QD
Sbjct 404 PDAQFDAAMKILQDAL 419
> mmu:72947 Agxt2l2, 2900006B13Rik; alanine-glyoxylate aminotransferase
2-like 2; K14286 alanine-glyoxylate aminotransferase
2-like [EC:2.6.1.-]
Length=467
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 23/86 (26%), Positives = 41/86 (47%), Gaps = 11/86 (12%)
Query 3 KEKCNLEWIRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCR---PTRGN 52
++K I D+RG GLF V++ ++ EA L LK +L P + N
Sbjct 349 QQKAKHPIIGDVRGTGLFIGVDLIKDETLRTPATEEAEYLVSRLKENYILLSIDGPGK-N 407
Query 53 VLRFLPPLCIDEAQMKDALQRIQDVF 78
+L+F PP+C + + + ++ D+
Sbjct 408 ILKFKPPMCFNVDNAQHVVAKLDDIL 433
> eco:b2662 gabT, ECK2656, JW2637; 4-aminobutyrate aminotransferase,
PLP-dependent (EC:2.6.1.19); K07250 4-aminobutyrate aminotransferase
/ (S)-3-amino-2-methylpropionate transaminase
[EC:2.6.1.19 2.6.1.22]
Length=426
Score = 34.3 bits (77), Expect = 0.096, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 41/78 (52%), Gaps = 11/78 (14%)
Query 11 IRDIRGRGLFNAVEI--NGSSGE-----AMQLCIELKNEGVL---CRPTRGNVLRFLPPL 60
I D+RG G A+E+ +G + ++ +++G++ C P NVLR L PL
Sbjct 345 IGDVRGLGAMIAIELFEDGDHNKPDAKLTAEIVARARDKGLILLSCGPYY-NVLRILVPL 403
Query 61 CIDEAQMKDALQRIQDVF 78
I++AQ++ L+ I F
Sbjct 404 TIEDAQIRQGLEIISQCF 421
> eco:b3359 argD, alaB, arg1, argG, dapC, dtu, ECK3347, JW3322;
bifunctional acetylornithine aminotransferase/succinyldiaminopimelate
aminotransferase (EC:2.6.1.11 2.6.1.17); K00821
acetylornithine/N-succinyldiaminopimelate aminotransferase [EC:2.6.1.11
2.6.1.17]
Length=406
Score = 34.3 bits (77), Expect = 0.100, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 1/62 (1%)
Query 13 DIRGRGLFNAVEINGS-SGEAMQLCIELKNEGVLCRPTRGNVLRFLPPLCIDEAQMKDAL 71
DIRG GL E+ G A GV+ +V+RF P L +++A + + +
Sbjct 334 DIRGMGLLIGAELKPQYKGRARDFLYAGAEAGVMVLNAGPDVMRFAPSLVVEDADIDEGM 393
Query 72 QR 73
QR
Sbjct 394 QR 395
> ath:AT4G39660 AGT2; AGT2 (ALANINE:GLYOXYLATE AMINOTRANSFERASE
2); alanine-glyoxylate transaminase/ catalytic/ pyridoxal
phosphate binding / transaminase; K00827 alanine-glyoxylate
transaminase / (R)-3-amino-2-methylpropionate-pyruvate transaminase
[EC:2.6.1.44 2.6.1.40]
Length=476
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 25/73 (34%), Positives = 35/73 (47%), Gaps = 12/73 (16%)
Query 11 IRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCRP--TRGNVLRFLPPLC 61
I D+RGRGL +E+ + E L +L+ G+L GNV R PP+C
Sbjct 397 IGDVRGRGLMVGIELVSDRKDKTPAKAETSVLFEQLRELGILVGKGGLHGNVFRIKPPMC 456
Query 62 I--DEAQ-MKDAL 71
D+A + DAL
Sbjct 457 FTKDDADFLVDAL 469
> hsa:85007 AGXT2L2, MGC117348, MGC15875, MGC45484; alanine-glyoxylate
aminotransferase 2-like 2; K14286 alanine-glyoxylate
aminotransferase 2-like [EC:2.6.1.-]
Length=450
Score = 32.3 bits (72), Expect = 0.37, Method: Composition-based stats.
Identities = 22/78 (28%), Positives = 36/78 (46%), Gaps = 11/78 (14%)
Query 11 IRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCR---PTRGNVLRFLPPL 60
+ D+RG GLF V++ ++ EA L LK VL P R N+L+F PP+
Sbjct 357 VGDVRGVGLFIGVDLIKDEATRTPATEEAAYLVSRLKENYVLLSTDGPGR-NILKFKPPM 415
Query 61 CIDEAQMKDALQRIQDVF 78
C + + ++ +
Sbjct 416 CFSLDNARQVVAKLDAIL 433
> dre:619269 agxt2, MGC114195, im:7153274, zgc:114195; alanine-glyoxylate
aminotransferase 2; K00827 alanine-glyoxylate transaminase
/ (R)-3-amino-2-methylpropionate-pyruvate transaminase
[EC:2.6.1.44 2.6.1.40]
Length=517
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 33/75 (44%), Gaps = 12/75 (16%)
Query 1 KMKEKCNLEWIRDIRGRGLFNAVEINGSSGEAMQLCIELKNE--------GVLCRP--TR 50
K+++K E I D+RG+GL VE+ L E NE GVL
Sbjct 425 KLRDK--YEIIGDVRGKGLQIGVEMVKDKASRTPLPQEAMNEIFEDTKDMGVLIGKGGLY 482
Query 51 GNVLRFLPPLCIDEA 65
G R PP+CI +A
Sbjct 483 GQTFRIKPPMCITKA 497
> tgo:TGME49_073800 hypothetical protein
Length=3633
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 22/56 (39%), Positives = 25/56 (44%), Gaps = 4/56 (7%)
Query 9 EWIRDIRGRGLFNAVEINGSSGEAMQLCIELKNEGV----LCRPTRGNVLRFLPPL 60
EW IRGRG A E GSS E+ L E V L + G + FL PL
Sbjct 951 EWTLSIRGRGSSKASESRGSSIESGGLGTHFSCENVEAVLLAWTSDGALRAFLLPL 1006
> cel:T01B11.2 hypothetical protein
Length=467
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 36/80 (45%), Gaps = 10/80 (12%)
Query 9 EWIRDIRGRGLFNAVEI---NGSSGEAMQLCIEL-----KNEGVLCRPT--RGNVLRFLP 58
E I DIRG GLF +++ + +L I K+ G+L N+L+ P
Sbjct 383 ECIGDIRGVGLFWGIDLVKDRNTREPDQKLAIATILALRKSYGILLNADGPHTNILKIKP 442
Query 59 PLCIDEAQMKDALQRIQDVF 78
PLC +E + + + + V
Sbjct 443 PLCFNENNILETVTALDQVL 462
> dre:557205 MGC123007; zgc:123007; K14286 alanine-glyoxylate
aminotransferase 2-like [EC:2.6.1.-]
Length=447
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 11/64 (17%)
Query 11 IRDIRGRGLFNAVEI-------NGSSGEAMQLCIELKNEGVLCRPTRG---NVLRFLPPL 60
I D+RG GLF +E+ ++ EA L LK + ++ T G +V++F PP+
Sbjct 357 IGDVRGVGLFIGMELVKDRESREPATEEAAHLVRRLKEDRIV-MSTDGPWDSVIKFKPPM 415
Query 61 CIDE 64
C +
Sbjct 416 CFSK 419
> sce:YPR184W GDB1; Gdb1p (EC:3.2.1.33 2.4.1.25); K01196 glycogen
debranching enzyme [EC:2.4.1.25 3.2.1.33]
Length=1536
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 22 AVEINGSSGEAMQLCIELKNEGVL 45
A+EING A++ IELKN+G+
Sbjct 1271 AIEINGLLKSALRFVIELKNKGLF 1294
> ath:AT4G36950 MAPKKK21; ATP binding / protein kinase/ protein
serine/threonine kinase
Length=336
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 4/43 (9%)
Query 8 LEWIR-DIRGRGLFNAVEI---NGSSGEAMQLCIELKNEGVLC 46
+EWIR + G G F+ V + +GSS +A + +K+ GV+C
Sbjct 1 MEWIRRETIGHGSFSTVSLATTSGSSSKAFPSLMAVKSSGVVC 43
> hsa:84171 LOXL4, FLJ21889, LOXC; lysyl oxidase-like 4; K00280
lysyl oxidase-like protein 2/3/4 [EC:1.4.3.-]
Length=756
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 12/41 (29%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 10 WIRDIRGRGLFNAVEINGSSGEAMQLCIELKNEGVLCRPTR 50
W+ ++R G ++++ GS+G + C ++ GV+C P R
Sbjct 96 WLDNVRCVGTESSLDQCGSNGWGVSDCSHSEDVGVICHPRR 136
Lambda K H
0.324 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044675024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40