bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
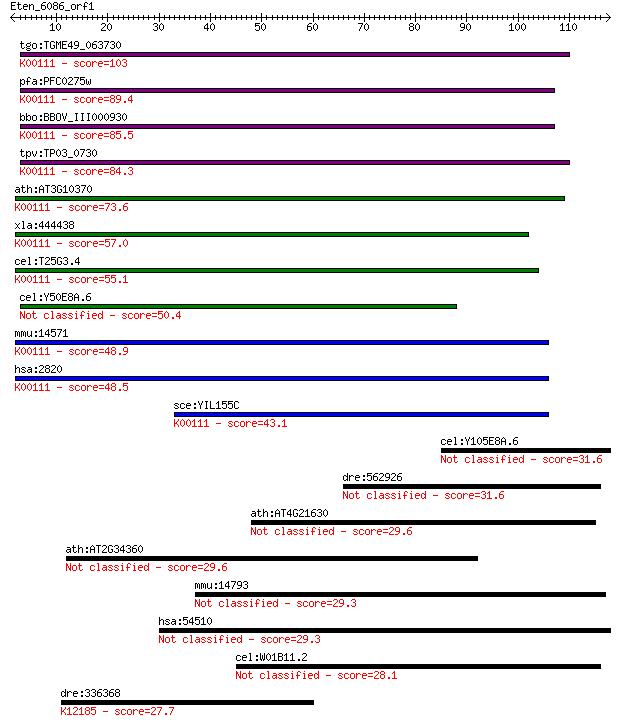
Query= Eten_6086_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_063730 glycerol-3-phosphate dehydrogenase, putative... 103 2e-22
pfa:PFC0275w FAD-dependent glycerol-3-phosphate dehydrogenase,... 89.4 2e-18
bbo:BBOV_III000930 17.m07109; FAD-dependent glycerol-3-phospha... 85.5 4e-17
tpv:TP03_0730 glycerol-3-phosphate dehydrogenase (EC:1.1.3.21)... 84.3 9e-17
ath:AT3G10370 SDP6; SDP6 (SUGAR-DEPENDENT 6); glycerol-3-phosp... 73.6 1e-13
xla:444438 gpd2, MGC83596; glycerol-3-phosphate dehydrogenase ... 57.0 2e-08
cel:T25G3.4 hypothetical protein; K00111 glycerol-3-phosphate ... 55.1 5e-08
cel:Y50E8A.6 hypothetical protein 50.4 1e-06
mmu:14571 Gpd2, AA408484, AI448216, AU021455, AW494132, GPDH, ... 48.9 4e-06
hsa:2820 GPD2, GDH2, GPDM, mGPDH; glycerol-3-phosphate dehydro... 48.5 5e-06
sce:YIL155C GUT2; Gut2p (EC:1.1.5.3); K00111 glycerol-3-phosph... 43.1 2e-04
cel:Y105E8A.6 unc-95; UNCoordinated family member (unc-95) 31.6 0.69
dre:562926 abhydrolase domain containing 9-like 31.6 0.72
ath:AT4G21630 subtilase family protein 29.6 2.3
ath:AT2G34360 antiporter/ drug transporter/ transporter 29.6 2.6
mmu:14793 Cdca3, 2410005A12Rik, C8, Grcc8, Tome-1; cell divisi... 29.3 3.0
hsa:54510 PCDH18, DKFZp434B0923, KIAA1562, PCDH68L; protocadhe... 29.3 3.6
cel:W01B11.2 sulp-6; SULfate Permease family member (sulp-6); ... 28.1 7.0
dre:336368 vps37a, fb08f03, fj42c03, wu:fb08f03, wu:fj42c03, z... 27.7 8.7
> tgo:TGME49_063730 glycerol-3-phosphate dehydrogenase, putative
(EC:1.1.3.21); K00111 glycerol-3-phosphate dehydrogenase
[EC:1.1.5.3]
Length=653
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 55/110 (50%), Positives = 72/110 (65%), Gaps = 3/110 (2%)
Query 3 HLVRHYGFMARDVVELAK-KESLMEPLV--EGLPYLKAEVLYACRYEQARSVSDILARRL 59
HLV YGF+AR V ++AK + L +PL+ E P+LKAEVLY RYE ARSVSD+LARR+
Sbjct 538 HLVESYGFLARSVCQIAKERPDLSQPLISDEAFPFLKAEVLYGVRYEMARSVSDVLARRM 597
Query 60 PILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETFTAAP 109
+ F+D ID V +M +EL WS+A KKK+EA + +TF P
Sbjct 598 RLAFVDVAKAEACIDEVARIMGEELKWSSAKLEKKKEEAAAFLKTFRCLP 647
> pfa:PFC0275w FAD-dependent glycerol-3-phosphate dehydrogenase,
putative; K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=653
Score = 89.4 bits (220), Expect = 2e-18, Method: Composition-based stats.
Identities = 38/104 (36%), Positives = 66/104 (63%), Gaps = 0/104 (0%)
Query 3 HLVRHYGFMARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRLPIL 62
HLV +YG+++ +V ELAK+ ++ + PY++AE++YA RYE A ++SDI+ RR +
Sbjct 545 HLVSNYGYLSENVCELAKELNMFNKIDPTKPYIEAEIIYATRYEFANTISDIIGRRFRLG 604
Query 63 FLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETFT 106
F+D ++ + ID + ++ EL W+ NK +EAK Y + +
Sbjct 605 FIDSQVSNKVIDKIAQLLKNELTWNTEQVNKNVKEAKDYINSLS 648
> bbo:BBOV_III000930 17.m07109; FAD-dependent glycerol-3-phosphate
dehydrogenase (EC:1.1.99.5); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=619
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 40/104 (38%), Positives = 63/104 (60%), Gaps = 0/104 (0%)
Query 3 HLVRHYGFMARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRLPIL 62
HLV YG+ +R+V ++ + +++P+ PYL+ EVLY R E A + DILARR I
Sbjct 512 HLVESYGYHSREVARMSAEAKMLKPIHPDFPYLQGEVLYGVRREFACTPLDILARRTRIA 571
Query 63 FLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETFT 106
FLD+K ++DTVC +M +EL WS++ + + +A + T
Sbjct 572 FLDNKAAAASLDTVCDIMGRELSWSSSRRAELRSQAVDFFNTMN 615
> tpv:TP03_0730 glycerol-3-phosphate dehydrogenase (EC:1.1.3.21);
K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=615
Score = 84.3 bits (207), Expect = 9e-17, Method: Composition-based stats.
Identities = 41/107 (38%), Positives = 63/107 (58%), Gaps = 0/107 (0%)
Query 3 HLVRHYGFMARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRLPIL 62
HLV YG+ + DV ++AK+ +L+ + E P+L EV+Y R E A + D+LARR +
Sbjct 509 HLVESYGYKSEDVCKVAKENNLLTKIHENYPFLLGEVVYGIRNELACTPVDVLARRTRLA 568
Query 63 FLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETFTAAP 109
FLD K ++++D V S+M KEL W+ T EA+ Y + + P
Sbjct 569 FLDAKAALQSLDKVVSVMEKELNWNPKTKKTLYLEAQDYFQQMSHVP 615
> ath:AT3G10370 SDP6; SDP6 (SUGAR-DEPENDENT 6); glycerol-3-phosphate
dehydrogenase (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=629
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 40/107 (37%), Positives = 60/107 (56%), Gaps = 0/107 (0%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRLPI 61
+HL YG MA V +A++E L + L G P+L+AEV Y R+E S D +ARR I
Sbjct 512 KHLSHAYGSMADRVATIAQEEGLGKRLAHGHPFLEAEVAYCARHEYCESAVDFIARRCRI 571
Query 62 LFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETFTAA 108
FLD A+ V ++A E W + ++ Q+AK++ ETF ++
Sbjct 572 AFLDTDAAARALQRVVEILASEHKWDKSRQKQELQKAKEFLETFKSS 618
> xla:444438 gpd2, MGC83596; glycerol-3-phosphate dehydrogenase
2 (mitochondrial) (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=725
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/105 (36%), Positives = 57/105 (54%), Gaps = 6/105 (5%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEP-----LVEGLPYLKAEVLYACRYEQARSVSDILA 56
+HL YG A +V +LAK P LV PY++AEV YA R E A + D+++
Sbjct 493 QHLAATYGDQAFEVAKLAKVTGKRWPIVGKRLVSEFPYIEAEVKYALR-EYACTAVDVIS 551
Query 57 RRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKY 101
RR + FL+ + EA+ + +MAKEL W ++ + AKK+
Sbjct 552 RRTRLAFLNVQAAEEALPRIVDIMAKELNWKEQRKKEELETAKKF 596
> cel:T25G3.4 hypothetical protein; K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=722
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 55/107 (51%), Gaps = 6/107 (5%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEPLVEG-----LPYLKAEVLYACRYEQARSVSDILA 56
+HL YG A V + K P+V PYL AEV YA R E A + D++A
Sbjct 497 QHLSNTYGDRAFVVARMCKMTGKRWPIVGQRLHPEFPYLDAEVRYAVR-EYACTAIDVIA 555
Query 57 RRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAE 103
RR+ + FL+ E + V +M +ELGWS+A + ++A+ + +
Sbjct 556 RRMRLAFLNTYAAHEVLPDVVRVMGQELGWSSAEQRAQLEKARTFID 602
> cel:Y50E8A.6 hypothetical protein
Length=609
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 50/90 (55%), Gaps = 6/90 (6%)
Query 3 HLVRHYGFMARDVVELAKKESLMEPLVE-----GLPYLKAEVLYACRYEQARSVSDILAR 57
HL + YG +V++L K P++ P+L+AEV YA + E AR +DILAR
Sbjct 486 HLCQTYGDKVYEVLKLCKSTGQKFPVIGHRLHPDFPFLEAEVRYAVK-EYARIPADILAR 544
Query 58 RLPILFLDHKLGVEAIDTVCSMMAKELGWS 87
R + LD + + + + ++MA+EL WS
Sbjct 545 RTRLSLLDARAARQVLPRIVAIMAEELKWS 574
> mmu:14571 Gpd2, AA408484, AI448216, AU021455, AW494132, GPDH,
Gdm1, Gpd-m, Gpdh-m, TISP38; glycerol phosphate dehydrogenase
2, mitochondrial (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=727
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 56/109 (51%), Gaps = 10/109 (9%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEP-----LVEGLPYLKAEVLYACRYEQARSVSDILA 56
+HL + YG A +V ++A P LV PY++AEV Y + E A + D+++
Sbjct 495 QHLAKTYGDKAFEVAKMASVTGKRWPVVGVRLVSEFPYIEAEVKYGIK-EYACTAVDMIS 553
Query 57 RRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETF 105
RR + FL+ + EA+ + +M +EL WS +KQE + A F
Sbjct 554 RRTRLAFLNVQAAEEALPRIVELMGRELNWSEL----RKQEELETATRF 598
> hsa:2820 GPD2, GDH2, GPDM, mGPDH; glycerol-3-phosphate dehydrogenase
2 (mitochondrial) (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=727
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 55/109 (50%), Gaps = 10/109 (9%)
Query 2 EHLVRHYGFMARDVVELAKKESLMEP-----LVEGLPYLKAEVLYACRYEQARSVSDILA 56
+HL YG A +V ++A P LV PY++AEV Y + E A + D+++
Sbjct 495 QHLAATYGDKAFEVAKMASVTGKRWPIVGVRLVSEFPYIEAEVKYGIK-EYACTAVDMIS 553
Query 57 RRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETF 105
RR + FL+ + EA+ + +M +EL W + KKQE + A F
Sbjct 554 RRTRLAFLNVQAAEEALPRIVELMGRELNWD----DYKKQEQLETARKF 598
> sce:YIL155C GUT2; Gut2p (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=649
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 34/73 (46%), Gaps = 0/73 (0%)
Query 33 PYLKAEVLYACRYEQARSVSDILARRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATAN 92
P+ E+ Y+ +YE R+ D L RR FLD K + A+ +M E WS
Sbjct 575 PFTIGELKYSMQYEYCRTPLDFLLRRTRFAFLDAKEALNAVHATVKVMGDEFNWSEKKRQ 634
Query 93 KKKQEAKKYAETF 105
+ ++ + +TF
Sbjct 635 WELEKTVNFIKTF 647
> cel:Y105E8A.6 unc-95; UNCoordinated family member (unc-95)
Length=350
Score = 31.6 bits (70), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 85 GWSAATANKKKQEAKKYAETFTAAPEEERPERP 117
G NKKK+E +Y T APEE+ P+RP
Sbjct 182 GRDGEDGNKKKKEEIEYTLRLTPAPEEQIPQRP 214
> dre:562926 abhydrolase domain containing 9-like
Length=482
Score = 31.6 bits (70), Expect = 0.72, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 30/51 (58%), Gaps = 2/51 (3%)
Query 66 HKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETFTAAPEE-ERPE 115
H + +E D V +++ + W ATA+K KQEA +T T +PE+ RP+
Sbjct 392 HMVMMECPDVVNTLLHEFFLWQPATASKPKQEAAP-TKTNTKSPEDTSRPK 441
> ath:AT4G21630 subtilase family protein
Length=772
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 2/69 (2%)
Query 48 ARSVSDILAR--RLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYAETF 105
A+ D+LAR +P +F D+++G + + + + + SAAT + K AE
Sbjct 451 AKKPDDLLARYNSIPYIFTDYEIGTHILQYIRTTRSPTVRISAATTLNGQPAMTKVAEFS 510
Query 106 TAAPEEERP 114
+ P P
Sbjct 511 SRGPNSVSP 519
> ath:AT2G34360 antiporter/ drug transporter/ transporter
Length=480
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 36/80 (45%), Gaps = 2/80 (2%)
Query 12 ARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRLPILFLDHKLGVE 71
A+ V + S++E ++ G + ++ Y V +A LPIL L H L +
Sbjct 321 AKLAVRVVLSFSIVESILVGTVLILIRKIWGFAYSSDPEVVSHVASMLPILALGHSL--D 378
Query 72 AIDTVCSMMAKELGWSAATA 91
+ TV S +A+ GW A
Sbjct 379 SFQTVLSGVARGCGWQKIGA 398
> mmu:14793 Cdca3, 2410005A12Rik, C8, Grcc8, Tome-1; cell division
cycle associated 3
Length=266
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 38/84 (45%), Gaps = 8/84 (9%)
Query 37 AEVLYACRYEQARSVSDILARRLPILFLDHKLGVEAIDTVCSMMAKELGWSAATANKK-- 94
+EVL E S L R P LF D L + D S + L WS A + K
Sbjct 93 SEVLETEASESISSPELALPRETP-LFYDLDL---SSDPQLSPEDQLLPWSQAELDPKQV 148
Query 95 --KQEAKKYAETFTAAPEEERPER 116
K+EAK+ AET A+ ++P R
Sbjct 149 FTKEEAKQSAETIAASQNSDKPSR 172
> hsa:54510 PCDH18, DKFZp434B0923, KIAA1562, PCDH68L; protocadherin
18
Length=1135
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 20/90 (22%), Positives = 42/90 (46%), Gaps = 2/90 (2%)
Query 30 EGLPYLKAEVLYACR-YEQARSVSDILARRLPILFLD-HKLGVEAIDTVCSMMAKELGWS 87
+G P L +VL C +E A SV+ + LD + + ++ +C+++ +
Sbjct 661 KGNPQLHTKVLLKCMIFEYAESVTSTAMTSVSQASLDVSMIIIISLGAICAVLLVIMVLF 720
Query 88 AATANKKKQEAKKYAETFTAAPEEERPERP 117
A N++K++ + Y + + P+RP
Sbjct 721 ATRCNREKKDTRSYNCRVAESTYQHHPKRP 750
> cel:W01B11.2 sulp-6; SULfate Permease family member (sulp-6);
K14453 solute carrier family 26, other
Length=823
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 29/73 (39%), Gaps = 2/73 (2%)
Query 45 YEQARSVSDILARRL--PILFLDHKLGVEAIDTVCSMMAKELGWSAATANKKKQEAKKYA 102
YE A + I R PI F + ++ V I C + + T NKKK K+
Sbjct 553 YETANDIPGIKIFRFDSPIYFGNSEMFVRKIHQACGLNPLIVRGELETENKKKDARKEKE 612
Query 103 ETFTAAPEEERPE 115
E P+E E
Sbjct 613 EEDAEIPKETEKE 625
> dre:336368 vps37a, fb08f03, fj42c03, wu:fb08f03, wu:fj42c03,
zgc:73244, zgc:77637; vacuolar protein sorting 37A; K12185
ESCRT-I complex subunit VPS37
Length=387
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 27/49 (55%), Gaps = 5/49 (10%)
Query 11 MARDVVELAKKESLMEPLVEGLPYLKAEVLYACRYEQARSVSDILARRL 59
+ +VE+AKK +EP +EG + E+L C+YEQ + + ++
Sbjct 262 LVNSIVEMAKKNLQLEPQLEG---KRQEML--CKYEQLTQMKSVFETKM 305
Lambda K H
0.318 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2032807080
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40